Mechanisms of Histone Deacetylase Inhibitor-Regulated Gene Expression in Cancer Cells
- PMID: 24512308
- PMCID: PMC4492771
- DOI: 10.1089/ars.2014.5863
Mechanisms of Histone Deacetylase Inhibitor-Regulated Gene Expression in Cancer Cells
Abstract
Significance: Class I and II histone deacetylase inhibitors (HDACis) are approved for the treatment of cutaneous T-cell lymphoma and are undergoing clinical trials as single agents, and in combination, for other hematological and solid tumors. Understanding their mechanisms of action is essential for their more effective clinical use, and broadening their clinical potential.
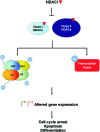
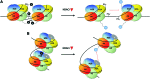
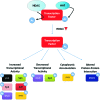
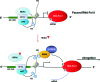
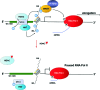
Recent advances: HDACi induce extensive transcriptional changes in tumor cells by activating and repressing similar numbers of genes. These transcriptional changes mediate, at least in part, HDACi-mediated growth inhibition, apoptosis, and differentiation. Here, we highlight two fundamental mechanisms by which HDACi regulate gene expression—histone and transcription factor acetylation. We also review the transcriptional responses invoked by HDACi, and compare these effects within and across tumor types.
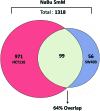
Critical issues: The mechanistic basis for how HDACi activate, and in particular repress gene expression, is not well understood. In addition, whether subsets of genes are reproducibly regulated by these agents both within and across tumor types has not been systematically addressed. A detailed understanding of the transcriptional changes elicited by HDACi in various tumor types, and the mechanistic basis for these effects, may provide insights into the specificity of these drugs for transformed cells and specific tumor types.
Future directions: Understanding the mechanisms by which HDACi regulate gene expression and an appreciation of their transcriptional targets could facilitate the ongoing clinical development of these emerging therapeutics. In particular, this knowledge could inform the design of rational drug combinations involving HDACi, and facilitate the identification of mechanism-based biomarkers of response.
Figures


Similar articles
-
DNA microarray profiling of genes differentially regulated by the histone deacetylase inhibitors vorinostat and LBH589 in colon cancer cell lines.BMC Med Genomics. 2009 Nov 30;2:67. doi: 10.1186/1755-8794-2-67. BMC Med Genomics. 2009. PMID: 19948057 Free PMC article.
-
ATF3 Repression of BCL-XL Determines Apoptotic Sensitivity to HDAC Inhibitors across Tumor Types.Clin Cancer Res. 2017 Sep 15;23(18):5573-5584. doi: 10.1158/1078-0432.CCR-17-0466. Epub 2017 Jun 13. Clin Cancer Res. 2017. PMID: 28611196 Free PMC article.
-
Apoptotic sensitivity of colon cancer cells to histone deacetylase inhibitors is mediated by an Sp1/Sp3-activated transcriptional program involving immediate-early gene induction.Cancer Res. 2010 Jan 15;70(2):609-20. doi: 10.1158/0008-5472.CAN-09-2327. Epub 2010 Jan 12. Cancer Res. 2010. PMID: 20068171 Free PMC article.
-
Histone deacetylase inhibitor (HDACI) mechanisms of action: emerging insights.Pharmacol Ther. 2014 Sep;143(3):323-36. doi: 10.1016/j.pharmthera.2014.04.004. Epub 2014 Apr 24. Pharmacol Ther. 2014. PMID: 24769080 Free PMC article. Review.
-
Enhancing the apoptotic and therapeutic effects of HDAC inhibitors.Cancer Lett. 2009 Aug 8;280(2):125-33. doi: 10.1016/j.canlet.2009.02.042. Epub 2009 Apr 8. Cancer Lett. 2009. PMID: 19359091 Review.
Cited by
-
Epigenetic mechanisms differentially regulate blood pressure and renal dysfunction in male and female Npr1 haplotype mice.FASEB J. 2024 Aug 15;38(15):e23858. doi: 10.1096/fj.202400714R. FASEB J. 2024. PMID: 39109516 Free PMC article.
-
Latency Reversing Agents: Kick and Kill of HTLV-1?Int J Mol Sci. 2021 May 24;22(11):5545. doi: 10.3390/ijms22115545. Int J Mol Sci. 2021. PMID: 34073995 Free PMC article. Review.
-
A Novel View of the Adult Stem Cell Compartment From the Perspective of a Quiescent Population of Very Small Embryonic-Like Stem Cells.Circ Res. 2017 Jan 6;120(1):166-178. doi: 10.1161/CIRCRESAHA.116.309362. Circ Res. 2017. PMID: 28057792 Free PMC article. Review.
-
Efficacy of HDAC Inhibitors in Driving Peroxisomal β-Oxidation and Immune Responses in Human Macrophages: Implications for Neuroinflammatory Disorders.Biomolecules. 2023 Nov 23;13(12):1696. doi: 10.3390/biom13121696. Biomolecules. 2023. PMID: 38136568 Free PMC article.
-
Enhanced Solubility, Permeability and Anticancer Activity of Vorinostat Using Tailored Mesoporous Silica Nanoparticles.Pharmaceutics. 2018 Dec 17;10(4):283. doi: 10.3390/pharmaceutics10040283. Pharmaceutics. 2018. PMID: 30562958 Free PMC article.
References
-
- Agricola E, Randall RA, Gaarenstroom T, Dupont S, and Hill CS. Recruitment of TIF1gamma to chromatin via its PHD finger-bromodomain activates its ubiquitin ligase and transcriptional repressor activities. Mol Cell 43: 85–96, 2011 - PubMed
-
- Allfrey VG. Post-synthetic modifications of histone structure: a mechanism for the control of chromosome structure by the modulation of histone-DNA interactions. In: Chromatin and Chromosome Structure, edited by Li HJ. and Eckhardt RE. New York: Academic, 1977, pp. 167–191
-
- Amin MR, Dudeja PK, Ramaswamy K, and Malakooti J. Involvement of Sp1 and Sp3 in differential regulation of human NHE3 promoter activity by sodium butyrate and IFN-gamma/TNF-alpha. Am J Physiol Gastrointest Liver Physiol 293: G374–G382, 2007 - PubMed
-
- Ammanamanchi S, Freeman JW, and Brattain MG. Acetylated sp3 is a transcriptional activator. J Biol Chem 278: 35775–35780, 2003 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

