A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events
- PMID: 24498085
- PMCID: PMC3909098
- DOI: 10.1371/journal.pone.0087361
A pan-cancer analysis of transcriptome changes associated with somatic mutations in U2AF1 reveals commonly altered splicing events
Abstract
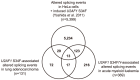
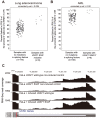
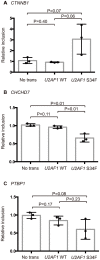
Although recurrent somatic mutations in the splicing factor U2AF1 (also known as U2AF35) have been identified in multiple cancer types, the effects of these mutations on the cancer transcriptome have yet to be fully elucidated. Here, we identified splicing alterations associated with U2AF1 mutations across distinct cancers using DNA and RNA sequencing data from The Cancer Genome Atlas (TCGA). Using RNA-Seq data from 182 lung adenocarcinomas and 167 acute myeloid leukemias (AML), in which U2AF1 is somatically mutated in 3-4% of cases, we identified 131 and 369 splicing alterations, respectively, that were significantly associated with U2AF1 mutation. Of these, 30 splicing alterations were statistically significant in both lung adenocarcinoma and AML, including three genes in the Cancer Gene Census, CTNNB1, CHCHD7, and PICALM. Cell line experiments expressing U2AF1 S34F in HeLa cells and in 293T cells provide further support that these altered splicing events are caused by U2AF1 mutation. Consistent with the function of U2AF1 in 3' splice site recognition, we found that S34F/Y mutations cause preferences for CAG over UAG 3' splice site sequences. This report demonstrates consistent effects of U2AF1 mutation on splicing in distinct cancer cell types.
Conflict of interest statement
Figures


Similar articles
-
Mutant U2AF1 Expression Alters Hematopoiesis and Pre-mRNA Splicing In Vivo.Cancer Cell. 2015 May 11;27(5):631-43. doi: 10.1016/j.ccell.2015.04.008. Cancer Cell. 2015. PMID: 25965570 Free PMC article.
-
Wild-Type U2AF1 Antagonizes the Splicing Program Characteristic of U2AF1-Mutant Tumors and Is Required for Cell Survival.PLoS Genet. 2016 Oct 24;12(10):e1006384. doi: 10.1371/journal.pgen.1006384. eCollection 2016 Oct. PLoS Genet. 2016. PMID: 27776121 Free PMC article.
-
U2AF1 mutations alter splice site recognition in hematological malignancies.Genome Res. 2015 Jan;25(1):14-26. doi: 10.1101/gr.181016.114. Epub 2014 Sep 29. Genome Res. 2015. PMID: 25267526 Free PMC article.
-
Splicing factor mutations and cancer.Wiley Interdiscip Rev RNA. 2014 Jul-Aug;5(4):445-59. doi: 10.1002/wrna.1222. Epub 2014 Feb 12. Wiley Interdiscip Rev RNA. 2014. PMID: 24523246 Review.
-
U2AF1 in various neoplastic diseases and relevant targeted therapies for malignant cancers with complex mutations (Review).Oncol Rep. 2024 Jan;51(1):5. doi: 10.3892/or.2023.8664. Epub 2023 Nov 17. Oncol Rep. 2024. PMID: 37975232 Free PMC article. Review.
Cited by
-
Changing the landscape of non-small cell lung cancer disparities.J Cancer Biol. 2021;2(2):33-38. doi: 10.46439/cancerbiology.2.020. J Cancer Biol. 2021. PMID: 35929605 Free PMC article.
-
Full-length transcript alterations in human bronchial epithelial cells with U2AF1 S34F mutations.Life Sci Alliance. 2023 Jul 24;6(10):e202000641. doi: 10.26508/lsa.202000641. Print 2023 Oct. Life Sci Alliance. 2023. PMID: 37487637 Free PMC article.
-
Investigating Open Reading Frames in Known and Novel Transcripts using ORFanage.bioRxiv [Preprint]. 2023 Mar 25:2023.03.23.533704. doi: 10.1101/2023.03.23.533704. bioRxiv. 2023. PMID: 36993373 Free PMC article. Updated. Preprint.
-
Defective control of pre-messenger RNA splicing in human disease.J Cell Biol. 2016 Jan 4;212(1):13-27. doi: 10.1083/jcb.201510032. J Cell Biol. 2016. PMID: 26728853 Free PMC article. Review.
-
Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells.Nat Commun. 2017 Jul 19;8:16027. doi: 10.1038/ncomms16027. Nat Commun. 2017. PMID: 28722025 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

