In vivo discovery of immunotherapy targets in the tumour microenvironment
- PMID: 24476824
- PMCID: PMC4052214
- DOI: 10.1038/nature12988
In vivo discovery of immunotherapy targets in the tumour microenvironment
Abstract
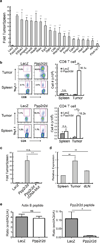
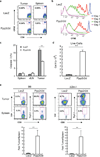
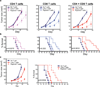
Recent clinical trials showed that targeting of inhibitory receptors on T cells induces durable responses in a subset of cancer patients, despite advanced disease. However, the regulatory switches controlling T-cell function in immunosuppressive tumours are not well understood. Here we show that such inhibitory mechanisms can be systematically discovered in the tumour microenvironment. We devised an in vivo pooled short hairpin RNA (shRNA) screen in which shRNAs targeting negative regulators became highly enriched in murine tumours by releasing a block on T-cell proliferation upon tumour antigen recognition. Such shRNAs were identified by deep sequencing of the shRNA cassette from T cells infiltrating tumour or control tissues. One of the target genes was Ppp2r2d, a regulatory subunit of the PP2A phosphatase family. In tumours, Ppp2r2d knockdown inhibited T-cell apoptosis and enhanced T-cell proliferation as well as cytokine production. Key regulators of immune function can therefore be discovered in relevant tissue microenvironments.
Figures

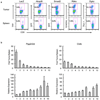



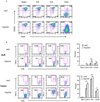
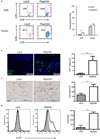
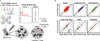

Comment in
-
Cancer: interference identifies immune modulators.Nature. 2014 Feb 6;506(7486):39-40. doi: 10.1038/nature13050. Epub 2014 Jan 29. Nature. 2014. PMID: 24476819 No abstract available.
-
Cancer: Live screening of immunotherapy targets.Nat Rev Drug Discov. 2014 Apr;13(4):258. doi: 10.1038/nrd4289. Epub 2014 Mar 21. Nat Rev Drug Discov. 2014. PMID: 24658309 No abstract available.
Similar articles
-
Checkpoint blockade immunotherapy enhances the frequency and effector function of murine tumor-infiltrating T cells but does not alter TCRβ diversity.Cancer Immunol Immunother. 2019 Jul;68(7):1095-1106. doi: 10.1007/s00262-019-02346-4. Epub 2019 May 18. Cancer Immunol Immunother. 2019. PMID: 31104075 Free PMC article.
-
Inhibition of melanoma growth after treatment with dendritic cells in a Tyr-SV40E murine model requires CD4+ T cells but not CD8+ T cells.Melanoma Res. 2004 Dec;14(6):555-62. doi: 10.1097/00008390-200412000-00019. Melanoma Res. 2004. PMID: 15577330
-
Topical treatment of all-trans retinoic acid inhibits murine melanoma partly by promoting CD8+ T-cell immunity.Immunology. 2017 Oct;152(2):287-297. doi: 10.1111/imm.12768. Epub 2017 Jun 29. Immunology. 2017. PMID: 28556970 Free PMC article.
-
Immuno-oncology: understanding the function and dysfunction of the immune system in cancer.Ann Oncol. 2012 Sep;23 Suppl 8(Suppl 8):viii6-9. doi: 10.1093/annonc/mds256. Ann Oncol. 2012. PMID: 22918931 Free PMC article. Review.
-
T-cell programming in pancreatic adenocarcinoma: a review.Cancer Gene Ther. 2017 Mar;24(3):106-113. doi: 10.1038/cgt.2016.66. Epub 2016 Dec 2. Cancer Gene Ther. 2017. PMID: 27910859 Review.
Cited by
-
Novel macrophage-related gene prognostic index for glioblastoma associated with M2 macrophages and T cell dysfunction.Front Immunol. 2022 Sep 13;13:941556. doi: 10.3389/fimmu.2022.941556. eCollection 2022. Front Immunol. 2022. PMID: 36177003 Free PMC article.
-
ACLY and ACSS2 link nutrient-dependent chromatin accessibility to CD8 T cell effector responses.J Exp Med. 2024 Sep 2;221(9):e20231820. doi: 10.1084/jem.20231820. Epub 2024 Aug 16. J Exp Med. 2024. PMID: 39150482
-
Fatty acid synthase (FASN) is a tumor-cell-intrinsic metabolic checkpoint restricting T-cell immunity.Cell Death Discov. 2024 Sep 30;10(1):417. doi: 10.1038/s41420-024-02184-z. Cell Death Discov. 2024. PMID: 39349429 Free PMC article.
-
Impaired CXCL12 signaling contributes to resistance of pancreatic cancer subpopulations to T cell-mediated cytotoxicity.Oncoimmunology. 2022 Feb 3;11(1):2027136. doi: 10.1080/2162402X.2022.2027136. eCollection 2022. Oncoimmunology. 2022. PMID: 35127250 Free PMC article.
-
Humanized Mouse Model Mimicking Pathology of Human Tuberculosis for in vivo Evaluation of Drug Regimens.Front Immunol. 2019 Jan 31;10:89. doi: 10.3389/fimmu.2019.00089. eCollection 2019. Front Immunol. 2019. PMID: 30766535 Free PMC article.
References
-
- Galon J, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313:1960–1964. - PubMed
-
- Mahmoud SM, et al. Tumor-Infiltrating CD8+ Lymphocytes Predict Clinical Outcome in Breast Cancer. J Clin Oncol. 2011;29:1949–1955. - PubMed
-
- Bindea G, et al. Spatiotemporal dynamics of intratumoral immune cells reveal the immune landscape in human cancer. Immunity. 2013;39:782–795. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

