H2B ubiquitylation modulates spliceosome assembly and function in budding yeast
- PMID: 24476359
- PMCID: PMC3989899
- DOI: 10.1111/boc.201400003
H2B ubiquitylation modulates spliceosome assembly and function in budding yeast
Abstract
Background information: Commitment to splicing occurs co-transcriptionally, but a major unanswered question is the extent to which various modifications of chromatin, the template for transcription in vivo, contribute to the regulation of splicing.
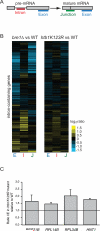
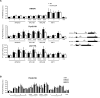
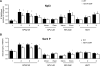
Results: Here, we perform genome-wide analyses showing that inhibition of specific marks - H2B ubiquitylation, H3K4 methylation and H3K36 methylation - perturbs splicing in budding yeast, with each modification exerting gene-specific effects. Furthermore, semi-quantitative mass spectrometry on purified nuclear mRNPs and chromatin immunoprecipitation analysis on intron-containing genes indicated that H2B ubiquitylation, but not Set1-, Set2- or Dot1-dependent H3 methylation, stimulates recruitment of the early splicing factors, namely U1 and U2 snRNPs, onto nascent RNAs.
Conclusions: These results suggest that histone modifications impact splicing of distinct subsets of genes using distinct pathways.
Keywords: H2B ubiquitylation; Histone marks; Pre-mRNA Splicing; snRNP.
© 2014 Société Française des Microscopies and Société de Biologie Cellulaire de France. Published by John Wiley & Sons Ltd.
Figures


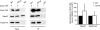
Similar articles
-
H3K36 Methylation and the Chromodomain Protein Eaf3 Are Required for Proper Cotranscriptional Spliceosome Assembly.Cell Rep. 2019 Jun 25;27(13):3760-3769.e4. doi: 10.1016/j.celrep.2019.05.100. Cell Rep. 2019. PMID: 31242410 Free PMC article.
-
The histone variant H2A.Z promotes efficient cotranscriptional splicing in S. cerevisiae.Genes Dev. 2017 Apr 1;31(7):702-717. doi: 10.1101/gad.295188.116. Genes Dev. 2017. PMID: 28446598 Free PMC article.
-
Cotranscriptional spliceosome assembly dynamics and the role of U1 snRNA:5'ss base pairing in yeast.Mol Cell. 2005 Jul 1;19(1):65-75. doi: 10.1016/j.molcel.2005.05.006. Mol Cell. 2005. PMID: 15989965
-
Histone H2B ubiquitylation: Connections to transcription and effects on chromatin structure.Biochim Biophys Acta Gene Regul Mech. 2024 Jun;1867(2):195018. doi: 10.1016/j.bbagrm.2024.195018. Epub 2024 Feb 6. Biochim Biophys Acta Gene Regul Mech. 2024. PMID: 38331024 Review.
-
A genetic and molecular toolbox for analyzing histone ubiquitylation and sumoylation in yeast.Methods. 2011 Jul;54(3):296-303. doi: 10.1016/j.ymeth.2011.02.003. Epub 2011 Feb 15. Methods. 2011. PMID: 21310241 Free PMC article. Review.
Cited by
-
Ready, SET, Go: Post-translational regulation of the histone lysine methylation network in budding yeast.J Biol Chem. 2021 Aug;297(2):100939. doi: 10.1016/j.jbc.2021.100939. Epub 2021 Jul 3. J Biol Chem. 2021. PMID: 34224729 Free PMC article. Review.
-
Nascent RNA and the Coordination of Splicing with Transcription.Cold Spring Harb Perspect Biol. 2019 Aug 1;11(8):a032227. doi: 10.1101/cshperspect.a032227. Cold Spring Harb Perspect Biol. 2019. PMID: 31371351 Free PMC article. Review.
-
The ubiquitin-selective chaperone Cdc48/p97 associates with Ubx3 to modulate monoubiquitylation of histone H2B.Nucleic Acids Res. 2014;42(17):10975-86. doi: 10.1093/nar/gku786. Epub 2014 Sep 2. Nucleic Acids Res. 2014. PMID: 25183520 Free PMC article.
-
Perfect timing: splicing and transcription rates in living cells.Wiley Interdiscip Rev RNA. 2017 Mar;8(2):10.1002/wrna.1401. doi: 10.1002/wrna.1401. Epub 2016 Nov 21. Wiley Interdiscip Rev RNA. 2017. PMID: 27873472 Free PMC article. Review.
-
Chromatin as a Platform for Modulating the Replication Stress Response.Genes (Basel). 2018 Dec 11;9(12):622. doi: 10.3390/genes9120622. Genes (Basel). 2018. PMID: 30544989 Free PMC article. Review.
References
-
- Alber F, Dokudovskaya S, Veenhoff LM, Zhang W, Kipper J, Devos D, Suprapto A, Karni-Schmidt O, Williams R, Chait BT, Sali A, Rout MP. The molecular architecture of the nuclear pore complex. Nature. 2007;450:695–701. - PubMed
-
- Babour A, Dargemont C, Stutz F. Ubiquitin and assembly of export competent mRNP. Biochim Biophys Acta. 2012;1819:521–530. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

