Composition of seed sequence is a major determinant of microRNA targeting patterns
- PMID: 24470575
- PMCID: PMC4016705
- DOI: 10.1093/bioinformatics/btu045
Composition of seed sequence is a major determinant of microRNA targeting patterns
Abstract
Motivation: MicroRNAs (miRNAs) are small non-coding RNAs that are extensively involved in gene expression regulation. One major roadblock in functional miRNA studies is the reliable prediction of genes targeted by miRNAs, as rules defining miRNA target recognition have not been well-established to date. Availability of high-throughput experimental data from a recent CLASH (cross linking, ligation and sequencing of hybrids) study has presented an unprecedented opportunity to characterize miRNA target recognition patterns, which may provide guidance for improved miRNA target prediction.
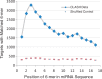
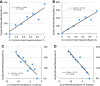
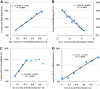
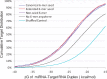
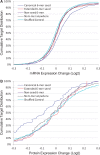
Results: The CLASH data were analysed to identify distinctive sequence features that characterize canonical and non-canonical miRNA target types. Most miRNA targets were of non-canonical type, i.e. without involving perfect pairing to canonical miRNA seed region. Different miRNAs have distinct targeting patterns, and this miRNA-to-miRNA variability was associated with seed sequence composition. Specifically, seed-based canonical target recognition was dependent on the GC content of the miRNA seed. For miRNAs with low GC content of the seed region, non-canonical targeting was the dominant mechanism for target recognition. In contrast to canonical targeting, non-canonical targeting did not lead to significant target downregulation at either the RNA or protein level.
Contact: xwang@radonc.wustl.edu.
Figures
Similar articles
-
Modified Cross-Linking, Ligation, and Sequencing of Hybrids (qCLASH) Identifies Kaposi's Sarcoma-Associated Herpesvirus MicroRNA Targets in Endothelial Cells.J Virol. 2018 Mar 28;92(8):e02138-17. doi: 10.1128/JVI.02138-17. Print 2018 Apr 15. J Virol. 2018. PMID: 29386283 Free PMC article.
-
Improving microRNA target prediction by modeling with unambiguously identified microRNA-target pairs from CLIP-ligation studies.Bioinformatics. 2016 May 1;32(9):1316-22. doi: 10.1093/bioinformatics/btw002. Epub 2016 Jan 6. Bioinformatics. 2016. PMID: 26743510 Free PMC article.
-
MicroRNA Target Recognition: Insights from Transcriptome-Wide Non-Canonical Interactions.Mol Cells. 2016 May 31;39(5):375-81. doi: 10.14348/molcells.2016.0013. Epub 2016 Apr 27. Mol Cells. 2016. PMID: 27117456 Free PMC article. Review.
-
Cross-Linking Ligation and Sequencing of Hybrids (qCLASH) Reveals an Unpredicted miRNA Targetome in Melanoma Cells.Cancers (Basel). 2021 Mar 4;13(5):1096. doi: 10.3390/cancers13051096. Cancers (Basel). 2021. PMID: 33806450 Free PMC article.
-
IsomiRs: Expanding the miRNA repression toolbox beyond the seed.Biochim Biophys Acta Gene Regul Mech. 2020 Apr;1863(4):194373. doi: 10.1016/j.bbagrm.2019.03.005. Epub 2019 Apr 4. Biochim Biophys Acta Gene Regul Mech. 2020. PMID: 30953728 Free PMC article. Review.
Cited by
-
A Thorough Navigation of miRNA's Blueprint in Crafting Cardiovascular Fate.Health Sci Rep. 2024 Nov 5;7(11):e70136. doi: 10.1002/hsr2.70136. eCollection 2024 Nov. Health Sci Rep. 2024. PMID: 39502130 Free PMC article.
-
Non-Canonical Targets of MicroRNAs: Role in Transcriptional Regulation, Disease Pathogenesis and Potential for Therapeutic Targets.Microrna. 2024;13(2):83-95. doi: 10.2174/0122115366278651240105071533. Microrna. 2024. PMID: 38317474 Review.
-
Computational Methods and Software Tools for Functional Analysis of miRNA Data.Biomolecules. 2020 Aug 28;10(9):1252. doi: 10.3390/biom10091252. Biomolecules. 2020. PMID: 32872205 Free PMC article. Review.
-
Novel miR-29b target regulation patterns are revealed in two different cell lines.Sci Rep. 2019 Nov 25;9(1):17449. doi: 10.1038/s41598-019-53868-x. Sci Rep. 2019. PMID: 31767948 Free PMC article.
-
Implications of SARS-CoV-2 Mutations for Genomic RNA Structure and Host microRNA Targeting.Int J Mol Sci. 2020 Jul 7;21(13):4807. doi: 10.3390/ijms21134807. Int J Mol Sci. 2020. PMID: 32645951 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

