Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits
- PMID: 24465181
- PMCID: PMC3897358
- DOI: 10.1371/journal.pbio.1001773
Conservation and divergence of regulatory strategies at Hox Loci and the origin of tetrapod digits
Abstract
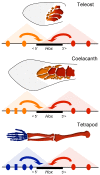
The evolution of tetrapod limbs from fish fins enabled the conquest of land by vertebrates and thus represents a key step in evolution. Despite the use of comparative gene expression analyses, critical aspects of this transformation remain controversial, in particular the origin of digits. Hoxa and Hoxd genes are essential for the specification of the different limb segments and their functional abrogation leads to large truncations of the appendages. Here we show that the selective transcription of mouse Hoxa genes in proximal and distal limbs is related to a bimodal higher order chromatin structure, similar to that reported for Hoxd genes, thus revealing a generic regulatory strategy implemented by both gene clusters during limb development. We found the same bimodal chromatin architecture in fish embryos, indicating that the regulatory mechanism used to pattern tetrapod limbs may predate the divergence between fish and tetrapods. However, when assessed in mice, both fish regulatory landscapes triggered transcription in proximal rather than distal limb territories, supporting an evolutionary scenario whereby digits arose as tetrapod novelties through genetic retrofitting of preexisting regulatory landscapes. We discuss the possibility to consider regulatory circuitries, rather than expression patterns, as essential parameters to define evolutionary synapomorphies.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

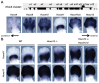
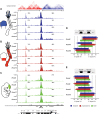

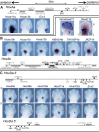
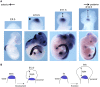
Comment in
-
A footnote to the evolution of digits.PLoS Biol. 2014 Jan;12(1):e1001774. doi: 10.1371/journal.pbio.1001774. Epub 2014 Jan 21. PLoS Biol. 2014. PMID: 24465182 Free PMC article. No abstract available.
Similar articles
-
Appendage expression driven by the Hoxd Global Control Region is an ancient gnathostome feature.Proc Natl Acad Sci U S A. 2011 Aug 2;108(31):12782-6. doi: 10.1073/pnas.1109993108. Epub 2011 Jul 15. Proc Natl Acad Sci U S A. 2011. PMID: 21765002 Free PMC article.
-
Digits and fin rays share common developmental histories.Nature. 2016 Sep 8;537(7619):225-228. doi: 10.1038/nature19322. Epub 2016 Aug 17. Nature. 2016. PMID: 27533041 Free PMC article.
-
Tri-phasic expression of posterior Hox genes during development of pectoral fins in zebrafish: implications for the evolution of vertebrate paired appendages.Dev Biol. 2008 Oct 1;322(1):220-33. doi: 10.1016/j.ydbio.2008.06.032. Epub 2008 Jul 3. Dev Biol. 2008. PMID: 18638469
-
Evolution of vertebrate appendicular structures: Insight from genetic and palaeontological data.Dev Dyn. 2011 May;240(5):1005-16. doi: 10.1002/dvdy.22572. Epub 2011 Feb 18. Dev Dyn. 2011. PMID: 21337665 Review.
-
Tinkering with constraints in the evolution of the vertebrate limb anterior-posterior polarity.Novartis Found Symp. 2007;284:130-7; discussion 138-41, 158-63. doi: 10.1002/9780470319390.ch9. Novartis Found Symp. 2007. PMID: 17710851 Review.
Cited by
-
Similarities and differences in the regulation of HoxD genes during chick and mouse limb development.PLoS Biol. 2018 Nov 26;16(11):e3000004. doi: 10.1371/journal.pbio.3000004. eCollection 2018 Nov. PLoS Biol. 2018. PMID: 30475793 Free PMC article.
-
Principles of genome folding into topologically associating domains.Sci Adv. 2019 Apr 10;5(4):eaaw1668. doi: 10.1126/sciadv.aaw1668. eCollection 2019 Apr. Sci Adv. 2019. PMID: 30989119 Free PMC article. Review.
-
Evolutionary parallelisms of pectoral and pelvic network-anatomy from fins to limbs.Sci Adv. 2019 May 8;5(5):eaau7459. doi: 10.1126/sciadv.aau7459. eCollection 2019 May. Sci Adv. 2019. PMID: 31086814 Free PMC article.
-
Nanoscale spatial organization of the HoxD gene cluster in distinct transcriptional states.Proc Natl Acad Sci U S A. 2015 Nov 10;112(45):13964-9. doi: 10.1073/pnas.1517972112. Epub 2015 Oct 26. Proc Natl Acad Sci U S A. 2015. PMID: 26504220 Free PMC article.
-
Transcriptional Trajectories in Mouse Limb Buds Reveal the Transition from Anterior-Posterior to Proximal-Distal Patterning at Early Limb Bud Stage.J Dev Biol. 2020 Dec 7;8(4):31. doi: 10.3390/jdb8040031. J Dev Biol. 2020. PMID: 33297480 Free PMC article.
References
-
- Woltering JM, Duboule D (2010) The origin of digits: expression patterns versus regulatory mechanisms. Dev Cell 18: 526–532. - PubMed
-
- Wagner GP, Chiu CH (2001) The tetrapod limb: a hypothesis on its origin. J Exp Zool 291: 226–240. - PubMed
-
- Gaffney ES (1979) Tetrapod monophyly: a phylogenetic analysis. Bull Carnegie Mus Nat Hist 13: 92–105.
-
- Holmgren N (1952) An embryological analysis of the mammalian carpus and its bearing upon the question of the origin of the tetrapod limb. Acta Zoologica 33: 1–115.
-
- Coates MI, Jeffery JE, Rut M (2002) Fins to limbs: what the fossils say. Evol Dev 4: 390–401. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

