Liquid biopsies: genotyping circulating tumor DNA
- PMID: 24449238
- PMCID: PMC4820760
- DOI: 10.1200/JCO.2012.45.2011
Liquid biopsies: genotyping circulating tumor DNA
Abstract
Genotyping tumor tissue in search of somatic genetic alterations for actionable information has become routine practice in clinical oncology. Although these sequence alterations are highly informative, sampling tumor tissue has significant inherent limitations; tumor tissue is a single snapshot in time, is subject to selection bias resulting from tumor heterogeneity, and can be difficult to obtain. Cell-free fragments of DNA are shed into the bloodstream by cells undergoing apoptosis or necrosis, and the load of circulating cell-free DNA (cfDNA) correlates with tumor staging and prognosis. Moreover, recent advances in the sensitivity and accuracy of DNA analysis have allowed for genotyping of cfDNA for somatic genomic alterations found in tumors. The ability to detect and quantify tumor mutations has proven effective in tracking tumor dynamics in real time as well as serving as a liquid biopsy that can be used for a variety of clinical and investigational applications not previously possible.
Conflict of interest statement
Authors' disclosures of potential conflicts of interest and author contributions are found at the end of this article.
Figures
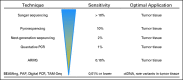

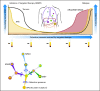
Similar articles
-
Circulating tumor DNA as a liquid biopsy for cancer.Clin Chem. 2015 Jan;61(1):112-23. doi: 10.1373/clinchem.2014.222679. Epub 2014 Nov 11. Clin Chem. 2015. PMID: 25388429 Review.
-
The Landscape of Actionable Genomic Alterations in Cell-Free Circulating Tumor DNA from 21,807 Advanced Cancer Patients.Clin Cancer Res. 2018 Aug 1;24(15):3528-3538. doi: 10.1158/1078-0432.CCR-17-3837. Epub 2018 May 18. Clin Cancer Res. 2018. PMID: 29776953
-
Prospective blinded study of somatic mutation detection in cell-free DNA utilizing a targeted 54-gene next generation sequencing panel in metastatic solid tumor patients.Oncotarget. 2015 Nov 24;6(37):40360-9. doi: 10.18632/oncotarget.5465. Oncotarget. 2015. PMID: 26452027 Free PMC article. Clinical Trial.
-
Circulating Cell-Free Tumor DNA Analysis of 50 Genes by Next-Generation Sequencing in the Prospective MOSCATO Trial.Clin Cancer Res. 2016 Jun 15;22(12):2960-8. doi: 10.1158/1078-0432.CCR-15-2470. Epub 2016 Jan 12. Clin Cancer Res. 2016. PMID: 26758560
-
Dynamic Treatment Stratification Using ctDNA.Recent Results Cancer Res. 2020;215:263-273. doi: 10.1007/978-3-030-26439-0_14. Recent Results Cancer Res. 2020. PMID: 31605234 Review.
Cited by
-
Predicting non-small cell lung cancer lymph node metastasis: integrating ctDNA mutation/methylation profiling with positron emission tomography-computed tomography (PET-CT) scan: protocol for a prospective clinical trial (LUNon-invasive Study).J Thorac Dis. 2024 Sep 30;16(9):6272-6285. doi: 10.21037/jtd-24-1033. Epub 2024 Sep 26. J Thorac Dis. 2024. PMID: 39444874 Free PMC article.
-
Circulating tumor-derived DNA is shorter than somatic DNA in plasma.Proc Natl Acad Sci U S A. 2015 Mar 17;112(11):3178-9. doi: 10.1073/pnas.1501321112. Epub 2015 Mar 2. Proc Natl Acad Sci U S A. 2015. PMID: 25733911 Free PMC article. No abstract available.
-
Diagnostic Accuracy of PIK3CA Mutation Detection by Circulating Free DNA in Breast Cancer: A Meta-Analysis of Diagnostic Test Accuracy.PLoS One. 2016 Jun 23;11(6):e0158143. doi: 10.1371/journal.pone.0158143. eCollection 2016. PLoS One. 2016. PMID: 27336598 Free PMC article.
-
A biocomposite-based rapid sampling assay for circulating cell-free DNA in liquid biopsy samples from human cancers.Sci Rep. 2020 Sep 10;10(1):14932. doi: 10.1038/s41598-020-72163-8. Sci Rep. 2020. PMID: 32913285 Free PMC article.
-
Circulating tumor DNA - A potential aid in the management of chordomas.Front Oncol. 2022 Oct 20;12:1016385. doi: 10.3389/fonc.2022.1016385. eCollection 2022. Front Oncol. 2022. PMID: 36338734 Free PMC article. Review.
References
-
- Mandel P, Metais P: Les acides nucleiques du plasma sanguin chez l'homme [in French] C R Seances Soc Biol Fil 142:241–243,1948 - PubMed
-
- Lo YM, Chiu RW: Genomic analysis of fetal nucleic acids in maternal blood Annu Rev Genomics Hum Genet 13:285–306,2012 - PubMed
-
- Lo YM Corbetta N Chamberlain PF, etal: Presence of fetal DNA in maternal plasma and serum Lancet 350:485–487,1997 - PubMed
-
- Lo YM Hjelm NM Fidler C, etal: Prenatal diagnosis of fetal RhD status by molecular analysis of maternal plasma N Engl J Med 339:1734–1738,1998 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

