Molecular control of δ-opioid receptor signalling
- PMID: 24413399
- PMCID: PMC3931418
- DOI: 10.1038/nature12944
Molecular control of δ-opioid receptor signalling
Abstract
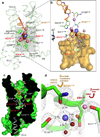
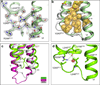
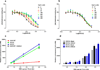
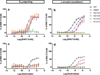
Opioids represent widely prescribed and abused medications, although their signal transduction mechanisms are not well understood. Here we present the 1.8 Å high-resolution crystal structure of the human δ-opioid receptor (δ-OR), revealing the presence and fundamental role of a sodium ion in mediating allosteric control of receptor functional selectivity and constitutive activity. The distinctive δ-OR sodium ion site architecture is centrally located in a polar interaction network in the seven-transmembrane bundle core, with the sodium ion stabilizing a reduced agonist affinity state, and thereby modulating signal transduction. Site-directed mutagenesis and functional studies reveal that changing the allosteric sodium site residue Asn 131 to an alanine or a valine augments constitutive β-arrestin-mediated signalling. Asp95Ala, Asn310Ala and Asn314Ala mutations transform classical δ-opioid antagonists such as naltrindole into potent β-arrestin-biased agonists. The data establish the molecular basis for allosteric sodium ion control in opioid signalling, revealing that sodium-coordinating residues act as 'efficacy switches' at a prototypic G-protein-coupled receptor.
Figures
Similar articles
-
Investigation of the selectivity of oxymorphone- and naltrexone-derived ligands via site-directed mutagenesis of opioid receptors: exploring the "address" recognition locus.J Med Chem. 2001 Mar 15;44(6):857-62. doi: 10.1021/jm000381r. J Med Chem. 2001. PMID: 11300867
-
Identification of selective agonists and positive allosteric modulators for µ- and δ-opioid receptors from a single high-throughput screen.J Biomol Screen. 2014 Oct;19(9):1255-65. doi: 10.1177/1087057114542975. Epub 2014 Jul 21. J Biomol Screen. 2014. PMID: 25047277
-
Selective delta-opioid receptor ligands: potential PET ligands based on naltrindole.Bioorg Med Chem Lett. 2001 Apr 9;11(7):939-43. doi: 10.1016/s0960-894x(01)00112-3. Bioorg Med Chem Lett. 2001. PMID: 11294396
-
The design of delta-selective opioid receptor antagonists.Farmaco. 1993 Feb;48(2):243-51. Farmaco. 1993. PMID: 8388215 Review.
-
Identifying ligand-specific signalling within biased responses: focus on δ opioid receptor ligands.Br J Pharmacol. 2015 Jan;172(2):435-48. doi: 10.1111/bph.12705. Epub 2014 Jul 1. Br J Pharmacol. 2015. PMID: 24665881 Free PMC article. Review.
Cited by
-
Structure of the adenosine A(2A) receptor bound to an engineered G protein.Nature. 2016 Aug 4;536(7614):104-7. doi: 10.1038/nature18966. Epub 2016 Jul 27. Nature. 2016. PMID: 27462812 Free PMC article.
-
Exploration of beta-arrestin isoform signaling pathways in delta opioid receptor agonist-induced convulsions.Front Pharmacol. 2022 Aug 11;13:914651. doi: 10.3389/fphar.2022.914651. eCollection 2022. Front Pharmacol. 2022. PMID: 36059958 Free PMC article.
-
Opioid receptors: Structural and mechanistic insights into pharmacology and signaling.Eur J Pharmacol. 2015 Sep 15;763(Pt B):206-13. doi: 10.1016/j.ejphar.2015.05.012. Epub 2015 May 14. Eur J Pharmacol. 2015. PMID: 25981301 Free PMC article. Review.
-
Mathematical analysis of the sodium sensitivity of the human histamine H3 receptor.In Silico Pharmacol. 2014 Dec;2(1):1. doi: 10.1186/s40203-014-0001-y. Epub 2014 May 24. In Silico Pharmacol. 2014. PMID: 27502620 Free PMC article.
-
Discovery of Novel Delta Opioid Receptor (DOR) Inverse Agonist and Irreversible (Non-Competitive) Antagonists.Molecules. 2021 Nov 5;26(21):6693. doi: 10.3390/molecules26216693. Molecules. 2021. PMID: 34771099 Free PMC article.
References
-
- Wootten D, Christopoulos A, Sexton PM. Emerging paradigms in GPCR allostery: implications for drug discovery. Nature reviews. Drug discovery. 2013;12:630–644. - PubMed
-
- Pert CB, Pasternak G, Snyder SH. Opiate agonists and antagonists discriminated by receptor binding in brain. Science. 1973;182:1359–1361. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

