Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct
- PMID: 24398669
- PMCID: PMC4076807
- DOI: 10.1093/carcin/bgu003
Molecular basis of aflatoxin-induced mutagenesis-role of the aflatoxin B1-formamidopyrimidine adduct
Abstract
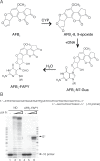
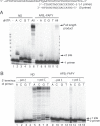
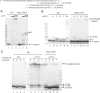
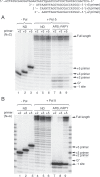
Aflatoxin B1 (AFB1) is a known carcinogen associated with early-onset hepatocellular carcinoma (HCC) and is thought to contribute to over half a million new HCCs per year. Although some of the fundamental risk factors are established, the molecular basis of AFB1-induced mutagenesis in primate cells has not been rigorously investigated. To gain insights into genome instability that is produced as a result of replicating DNAs containing AFB1 adducts, site-specific mutagenesis assays were used to establish the mutagenic potential of the persistent ring-opened AFB1 adduct, AFB1-formamidopyrimidine (AFB1-FAPY). This lesion was highly mutagenic, yielding replication error frequencies of 97%, with the predominant base substitution being a G to T transversion. This transversion is consistent with previous mutational data derived from aflatoxin-associated HCCs. In vitro translesion synthesis assays demonstrated that polymerase (pol) ζ was the most likely candidate polymerase that is responsible for the G to T mutations induced by this adduct.
© The Author 2014. Published by Oxford University Press. All rights reserved. For Permissions, please email: journals.permissions@oup.com.
Figures
Similar articles
-
Error-prone replication bypass of the primary aflatoxin B1 DNA adduct, AFB1-N7-Gua.J Biol Chem. 2014 Jun 27;289(26):18497-506. doi: 10.1074/jbc.M114.561563. Epub 2014 May 16. J Biol Chem. 2014. PMID: 24838242 Free PMC article.
-
DNA polymerase ζ limits chromosomal damage and promotes cell survival following aflatoxin exposure.Proc Natl Acad Sci U S A. 2016 Nov 29;113(48):13774-13779. doi: 10.1073/pnas.1609024113. Epub 2016 Nov 14. Proc Natl Acad Sci U S A. 2016. PMID: 27849610 Free PMC article.
-
Mutational properties of the primary aflatoxin B1-DNA adduct.Proc Natl Acad Sci U S A. 1996 Feb 20;93(4):1535-9. doi: 10.1073/pnas.93.4.1535. Proc Natl Acad Sci U S A. 1996. PMID: 8643667 Free PMC article.
-
Mechanisms underlying aflatoxin-associated mutagenesis - Implications in carcinogenesis.DNA Repair (Amst). 2019 May;77:76-86. doi: 10.1016/j.dnarep.2019.03.004. Epub 2019 Mar 7. DNA Repair (Amst). 2019. PMID: 30897375 Free PMC article. Review.
-
Mutations of the p53 tumor suppressor gene and ras oncogenes in aflatoxin hepatocarcinogenesis.Mutat Res. 1996 Oct;366(1):23-44. doi: 10.1016/s0165-1110(96)90005-6. Mutat Res. 1996. PMID: 8921985 Review.
Cited by
-
Eukaryotic DNA polymerase ζ.DNA Repair (Amst). 2015 May;29:47-55. doi: 10.1016/j.dnarep.2015.02.012. Epub 2015 Feb 19. DNA Repair (Amst). 2015. PMID: 25737057 Free PMC article. Review.
-
Recognition of DNA adducts by edited and unedited forms of DNA glycosylase NEIL1.DNA Repair (Amst). 2020 Jan;85:102741. doi: 10.1016/j.dnarep.2019.102741. Epub 2019 Nov 2. DNA Repair (Amst). 2020. PMID: 31733589 Free PMC article.
-
Functional analyses of single nucleotide polymorphic variants of the DNA glycosylase NEIL1 in sub-Saharan African populations.DNA Repair (Amst). 2023 Sep;129:103544. doi: 10.1016/j.dnarep.2023.103544. Epub 2023 Jul 20. DNA Repair (Amst). 2023. PMID: 37517321 Free PMC article.
-
Mutational spectra of aflatoxin B1 in vivo establish biomarkers of exposure for human hepatocellular carcinoma.Proc Natl Acad Sci U S A. 2017 Apr 11;114(15):E3101-E3109. doi: 10.1073/pnas.1700759114. Epub 2017 Mar 28. Proc Natl Acad Sci U S A. 2017. PMID: 28351974 Free PMC article.
-
Synthesis and Characterization of 15N5-Labeled Aflatoxin B1-Formamidopyrimidines and Aflatoxin B1-N7-Guanine from a Partial Double-Stranded Oligodeoxynucleotide as Internal Standards for Mass Spectrometric Measurements.ACS Omega. 2023 Apr 11;8(16):14841-14854. doi: 10.1021/acsomega.3c01328. eCollection 2023 Apr 25. ACS Omega. 2023. PMID: 37125130 Free PMC article.
References
-
- Ferlay J., et al. (2010). Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int. J. Cancer, 127, 2893–2917 - PubMed
-
- Baertschi S.W., et al. (1988). Preparation of the 8,9-epoxide of the mycotoxin aflatoxin B1: the ultimate carcinogenic species. J. Am. Chem. Soc., 110, 7929–7931
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

