SH2B1β interacts with STAT3 and enhances fibroblast growth factor 1-induced gene expression during neuronal differentiation
- PMID: 24396070
- PMCID: PMC3958036
- DOI: 10.1128/MCB.00940-13
SH2B1β interacts with STAT3 and enhances fibroblast growth factor 1-induced gene expression during neuronal differentiation
Abstract
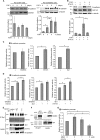
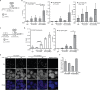
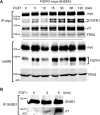
Neurite outgrowth is an essential process during neuronal differentiation as well as neuroregeneration. Thus, understanding the molecular and cellular control of neurite outgrowth will benefit patients with neurological diseases. We have previously shown that overexpression of the signaling adaptor protein SH2B1β promotes fibroblast growth factor 1 (FGF1)-induced neurite outgrowth (W. F. Lin, C. J. Chen, Y. J. Chang, S. L. Chen, I. M. Chiu, and L. Chen, Cell. Signal. 21:1060-1072, 2009). SH2B1β also undergoes nucleocytoplasmic shuttling and regulates a subset of neurotrophin-induced genes. Although these findings suggest that SH2B1β regulates gene expression, the nuclear role of SH2B1β was not known. In this study, we show that SH2B1β interacts with the transcription factor, signal transducer, and activator of transcription 3 (STAT3) in neuronal PC12 cells, cortical neurons, and COS7 fibroblasts. By affecting the subcellular distribution of STAT3, SH2B1β increased serine phosphorylation and the concomitant transcriptional activity of STAT3. As a result, overexpressing SH2B1β enhanced FGF1-induced expression of STAT3 target genes Egr1 and Cdh2. Chromatin immunoprecipitation assays further reveal that, in response to FGF1, overexpression of SH2B1β promotes the in vivo occupancy of STAT3-Sp1 heterodimers at the promoter of Egr1 and Cdh2. These findings establish a central role of SH2B1β in orchestrating signaling events to transcriptional activation through interacting and regulating STAT3-containing complexes during neuronal differentiation.
Figures


Similar articles
-
SH2B1beta enhances fibroblast growth factor 1 (FGF1)-induced neurite outgrowth through MEK-ERK1/2-STAT3-Egr1 pathway.Cell Signal. 2009 Jul;21(7):1060-72. doi: 10.1016/j.cellsig.2009.02.009. Epub 2009 Feb 26. Cell Signal. 2009. PMID: 19249349
-
Nucleocytoplasmic shuttling of the adapter protein SH2B1beta (SH2-Bbeta) is required for nerve growth factor (NGF)-dependent neurite outgrowth and enhancement of expression of a subset of NGF-responsive genes.Mol Endocrinol. 2009 Jul;23(7):1077-91. doi: 10.1210/me.2009-0011. Epub 2009 Apr 16. Mol Endocrinol. 2009. PMID: 19372237 Free PMC article.
-
New function of the adaptor protein SH2B1 in brain-derived neurotrophic factor-induced neurite outgrowth.PLoS One. 2013 Nov 15;8(11):e79619. doi: 10.1371/journal.pone.0079619. eCollection 2013. PLoS One. 2013. PMID: 24260264 Free PMC article.
-
SH2B1beta (SH2-Bbeta) enhances expression of a subset of nerve growth factor-regulated genes important for neuronal differentiation including genes encoding urokinase plasminogen activator receptor and matrix metalloproteinase 3/10.Mol Endocrinol. 2008 Feb;22(2):454-76. doi: 10.1210/me.2007-0384. Epub 2007 Oct 18. Mol Endocrinol. 2008. PMID: 17947375 Free PMC article.
-
Epigenetic regulation of Fgf1 transcription by CRTC1 and memory enhancement.Brain Res Bull. 2018 Jul;141:3-12. doi: 10.1016/j.brainresbull.2018.02.016. Epub 2018 Mar 2. Brain Res Bull. 2018. PMID: 29477835 Free PMC article. Review.
Cited by
-
SH2B1 orchestrates signaling events to filopodium formation during neurite outgrowth.Commun Integr Biol. 2015 Aug 31;8(4):e1044189. doi: 10.1080/19420889.2015.1044189. eCollection 2015 Jul-Aug. Commun Integr Biol. 2015. PMID: 26479731 Free PMC article.
-
A meta-analysis of 120 246 individuals identifies 18 new loci for fibrinogen concentration.Hum Mol Genet. 2016 Jan 15;25(2):358-70. doi: 10.1093/hmg/ddv454. Epub 2015 Nov 10. Hum Mol Genet. 2016. PMID: 26561523 Free PMC article.
-
STAT3 signal that mediates the neural plasticity is involved in willed-movement training in focal ischemic rats.J Zhejiang Univ Sci B. 2016 Jul;17(7):493-502. doi: 10.1631/jzus.B1500297. J Zhejiang Univ Sci B. 2016. PMID: 27381726 Free PMC article.
-
SH2B1 and IRSp53 proteins promote the formation of dendrites and dendritic branches.J Biol Chem. 2015 Mar 6;290(10):6010-21. doi: 10.1074/jbc.M114.603795. Epub 2015 Jan 13. J Biol Chem. 2015. PMID: 25586189 Free PMC article.
-
Takinib Inhibits Inflammation in Human Rheumatoid Arthritis Synovial Fibroblasts by Targeting the Janus Kinase-Signal Transducer and Activator of Transcription 3 (JAK/STAT3) Pathway.Int J Mol Sci. 2021 Nov 22;22(22):12580. doi: 10.3390/ijms222212580. Int J Mol Sci. 2021. PMID: 34830460 Free PMC article.
References
-
- Hashimoto M, Sagara Y, Langford D, Everall IP, Mallory M, Everson A, Digicaylioglu M, Masliah E. 2002. Fibroblast growth factor 1 regulates signaling via the glycogen synthase kinase-3beta pathway. Implications for neuroprotection. J. Biol. Chem. 277:32985–32991. 10.1074/jbc.M202803200 - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
