Structural polymorphism in the promoter of pfmrp2 confers Plasmodium falciparum tolerance to quinoline drugs
- PMID: 24372851
- PMCID: PMC4286016
- DOI: 10.1111/mmi.12505
Structural polymorphism in the promoter of pfmrp2 confers Plasmodium falciparum tolerance to quinoline drugs
Abstract
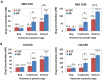
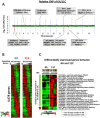
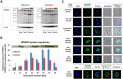
Drug resistance in Plasmodium falciparum remains a challenge for the malaria eradication programmes around the world. With the emergence of artemisinin resistance, the efficacy of the partner drugs in the artemisinin combination therapies (ACT) that include quinoline-based drugs is becoming critical. So far only few resistance markers have been identified from which only two transmembrane transporters namely PfMDR1 (an ATP-binding cassette transporter) and PfCRT (a drug-metabolite transporter) have been experimentally verified. Another P. falciparum transporter, the ATP-binding cassette containing multidrug resistance-associated protein (PfMRP2) represents an additional possible factor of drug resistance in P. falciparum. In this study, we identified a parasite clone that is derived from the 3D7 P. falciparum strain and shows increased resistance to chloroquine, mefloquine and quinine through the trophozoite and schizont stages. We demonstrate that the resistance phenotype is caused by a 4.1 kb deletion in the 5' upstream region of the pfmrp2 gene that leads to an alteration in the pfmrp2 transcription and thus increased level of PfMRP2 protein. These results also suggest the importance of putative promoter elements in regulation of gene expression during the P. falciparum intra-erythrocytic developmental cycle and the potential of genetic polymorphisms within these regions to underlie drug resistance.
© 2013 The Authors. Molecular Microbiology published by John Wiley & Sons Ltd.
Figures


Similar articles
-
Complex polymorphisms in the Plasmodium falciparum multidrug resistance protein 2 gene and its contribution to antimalarial response.Antimicrob Agents Chemother. 2014 Dec;58(12):7390-7. doi: 10.1128/AAC.03337-14. Epub 2014 Sep 29. Antimicrob Agents Chemother. 2014. PMID: 25267670 Free PMC article.
-
A Variant PfCRT Isoform Can Contribute to Plasmodium falciparum Resistance to the First-Line Partner Drug Piperaquine.mBio. 2017 May 9;8(3):e00303-17. doi: 10.1128/mBio.00303-17. mBio. 2017. PMID: 28487425 Free PMC article.
-
Inhibition of efflux of quinolines as new therapeutic strategy in malaria.Curr Top Med Chem. 2008;8(7):563-78. doi: 10.2174/156802608783955593. Curr Top Med Chem. 2008. PMID: 18473883 Review.
-
Mapping of the Plasmodium falciparum multidrug resistance gene 5'-upstream region, and evidence of induction of transcript levels by antimalarial drugs in chloroquine sensitive parasites.Mol Microbiol. 2003 Aug;49(3):671-83. doi: 10.1046/j.1365-2958.2003.03597.x. Mol Microbiol. 2003. PMID: 12864851
-
Molecular Mechanisms of Drug Resistance in Plasmodium falciparum Malaria.Annu Rev Microbiol. 2020 Sep 8;74:431-454. doi: 10.1146/annurev-micro-020518-115546. Annu Rev Microbiol. 2020. PMID: 32905757 Free PMC article. Review.
Cited by
-
Short tandem repeat polymorphism in the promoter region of cyclophilin 19B drives its transcriptional upregulation and contributes to drug resistance in the malaria parasite Plasmodium falciparum.PLoS Pathog. 2023 Jan 25;19(1):e1011118. doi: 10.1371/journal.ppat.1011118. eCollection 2023 Jan. PLoS Pathog. 2023. PMID: 36696458 Free PMC article.
-
Decreased susceptibility of Plasmodium falciparum to both dihydroartemisinin and lumefantrine in northern Uganda.Nat Commun. 2022 Oct 26;13(1):6353. doi: 10.1038/s41467-022-33873-x. Nat Commun. 2022. PMID: 36289202 Free PMC article.
-
A Plasmodium falciparum ATP-binding cassette transporter is essential for liver stage entry into schizogony.iScience. 2022 Apr 8;25(5):104224. doi: 10.1016/j.isci.2022.104224. eCollection 2022 May 20. iScience. 2022. PMID: 35521513 Free PMC article.
-
PMRT1, a Plasmodium-Specific Parasite Plasma Membrane Transporter, Is Essential for Asexual and Sexual Blood Stage Development.mBio. 2022 Apr 26;13(2):e0062322. doi: 10.1128/mbio.00623-22. Epub 2022 Apr 11. mBio. 2022. PMID: 35404116 Free PMC article.
-
Plasmodium falciparum Multidrug Resistance Proteins (pfMRPs).Front Pharmacol. 2021 Nov 1;12:759422. doi: 10.3389/fphar.2021.759422. eCollection 2021. Front Pharmacol. 2021. PMID: 34790129 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

