The machinery of macroautophagy
- PMID: 24366339
- PMCID: PMC3879710
- DOI: 10.1038/cr.2013.168
The machinery of macroautophagy
Abstract
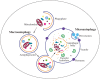
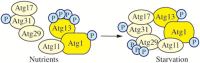
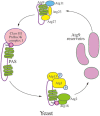
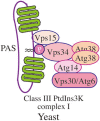
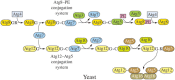
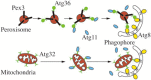
Autophagy is a primarily degradative pathway that takes place in all eukaryotic cells. It is used for recycling cytoplasm to generate macromolecular building blocks and energy under stress conditions, to remove superfluous and damaged organelles to adapt to changing nutrient conditions and to maintain cellular homeostasis. In addition, autophagy plays a critical role in cytoprotection by preventing the accumulation of toxic proteins and through its action in various aspects of immunity including the elimination of invasive microbes and its participation in antigen presentation. The most prevalent form of autophagy is macroautophagy, and during this process, the cell forms a double-membrane sequestering compartment termed the phagophore, which matures into an autophagosome. Following delivery to the vacuole or lysosome, the cargo is degraded and the resulting macromolecules are released back into the cytosol for reuse. The past two decades have resulted in a tremendous increase with regard to the molecular studies of autophagy being carried out in yeast and other eukaryotes. Part of the surge in interest in this topic is due to the connection of autophagy with a wide range of human pathophysiologies including cancer, myopathies, diabetes and neurodegenerative disease. However, there are still many aspects of autophagy that remain unclear, including the process of phagophore formation, the regulatory mechanisms that control its induction and the function of most of the autophagy-related proteins. In this review, we focus on macroautophagy, briefly describing the discovery of this process in mammalian cells, discussing the current views concerning the donor membrane that forms the phagophore, and characterizing the autophagy machinery including the available structural information.
Figures
Similar articles
-
How to control self-digestion: transcriptional, post-transcriptional, and post-translational regulation of autophagy.Trends Cell Biol. 2015 Jun;25(6):354-63. doi: 10.1016/j.tcb.2015.02.002. Epub 2015 Mar 8. Trends Cell Biol. 2015. PMID: 25759175 Free PMC article. Review.
-
Atg41/Icy2 regulates autophagosome formation.Autophagy. 2015;11(12):2288-99. doi: 10.1080/15548627.2015.1107692. Autophagy. 2015. PMID: 26565778 Free PMC article.
-
The world's first (and probably last) autophagy video game.Autophagy. 2023 Jan;19(1):352-357. doi: 10.1080/15548627.2022.2143212. Epub 2022 Nov 16. Autophagy. 2023. PMID: 36324276 Free PMC article.
-
Phagophore-lysosome/vacuole fusion in mutant yeast and mammalian cells.Autophagy. 2023 Sep;19(9):2595-2600. doi: 10.1080/15548627.2023.2205272. Epub 2023 Apr 28. Autophagy. 2023. PMID: 37083184 Free PMC article.
-
Autophagy in health and disease. 1. Regulation and significance of autophagy: an overview.Am J Physiol Cell Physiol. 2010 Apr;298(4):C776-85. doi: 10.1152/ajpcell.00507.2009. Epub 2010 Jan 20. Am J Physiol Cell Physiol. 2010. PMID: 20089931 Review.
Cited by
-
Ubiquitination regulates autophagy in cancer: simple modifications, promising targets.J Transl Med. 2024 Oct 31;22(1):985. doi: 10.1186/s12967-024-05565-1. J Transl Med. 2024. PMID: 39482684 Free PMC article. Review.
-
Exploring paraptosis as a therapeutic approach in cancer treatment.J Biomed Sci. 2024 Nov 4;31(1):101. doi: 10.1186/s12929-024-01089-4. J Biomed Sci. 2024. PMID: 39497143 Free PMC article. Review.
-
Spermidine induces autophagy by inhibiting the acetyltransferase EP300.Cell Death Differ. 2015 Mar;22(3):509-16. doi: 10.1038/cdd.2014.215. Epub 2014 Dec 19. Cell Death Differ. 2015. PMID: 25526088 Free PMC article.
-
A conserved mechanism of TOR-dependent RCK-mediated mRNA degradation regulates autophagy.Nat Cell Biol. 2015 Jul;17(7):930-942. doi: 10.1038/ncb3189. Epub 2015 Jun 22. Nat Cell Biol. 2015. PMID: 26098573 Free PMC article.
-
The Wnt Signaling Antagonist Dapper1 Accelerates Dishevelled2 Degradation via Promoting Its Ubiquitination and Aggregate-induced Autophagy.J Biol Chem. 2015 May 8;290(19):12346-54. doi: 10.1074/jbc.M115.654590. Epub 2015 Mar 30. J Biol Chem. 2015. PMID: 25825496 Free PMC article.
References
-
- Baba M, Osumi M, Ohsumi Y. Analysis of the membrane structures involved in autophagy in yeast by freeze-replica method. Cell Struct Funct. 1995;20:465–471. - PubMed
-
- Fimia GM, Stoykova A, Romagnoli A, et al. Ambra1 regulates autophagy and development of the nervous system. Nature. 2007;447:1121–1125. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

