Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin
- PMID: 24352422
- PMCID: PMC3877756
- DOI: 10.1101/gad.225888.113
Nucleosome-binding activities within JARID2 and EZH1 regulate the function of PRC2 on chromatin
Erratum in
- Genes Dev. 2014 Apr 15;28(8):921
Abstract
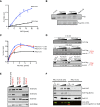
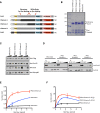
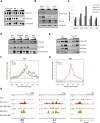
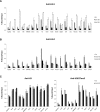
Polycomb-repressive complex 2 (PRC2) comprises specific members of the Polycomb group of epigenetic modulators. PRC2 catalyzes methylation of histone H3 at Lys 27 (H3K27me3) through its Enhancer of zeste (Ezh) constituent, of which there are two mammalian homologs: Ezh1 and Ezh2. Several ancillary factors, including Jarid2, modulate PRC2 function, with Jarid2 facilitating its recruitment to target genes. Jarid2, like Ezh2, is present in poorly differentiated and actively dividing cells, while Ezh1 associates with PRC2 in all cells, including resting cells. We found that Jarid2 exhibits nucleosome-binding activity that contributes to PRC2 stimulation. Moreover, such nucleosome-binding activity is exhibited by PRC2 comprising Ezh1 (PRC2-Ezh1), in contrast to PRC2-Ezh2. The presence of Ezh1 helps to maintain PRC2 occupancy on its target genes in myoblasts where Jarid2 is not expressed. Our findings allow us to propose a model in which PRC2-Ezh2 is important for the de novo establishment of H3K27me3 in dividing cells, whereas PRC2-Ezh1 is required for its maintenance in resting cells.
Keywords: EZH1; EZH2; JARID2; PRC2; Polycomb.
Figures



Similar articles
-
Jarid2 Methylation via the PRC2 Complex Regulates H3K27me3 Deposition during Cell Differentiation.Mol Cell. 2015 Mar 5;57(5):769-783. doi: 10.1016/j.molcel.2014.12.020. Epub 2015 Jan 22. Mol Cell. 2015. PMID: 25620564 Free PMC article.
-
Ezh1 and Ezh2 maintain repressive chromatin through different mechanisms.Mol Cell. 2008 Nov 21;32(4):503-18. doi: 10.1016/j.molcel.2008.11.004. Mol Cell. 2008. PMID: 19026781 Free PMC article.
-
Polycomb recruitment at the Class II transactivator gene.Mol Immunol. 2015 Oct;67(2 Pt B):482-91. doi: 10.1016/j.molimm.2015.08.003. Epub 2015 Aug 15. Mol Immunol. 2015. PMID: 26283540
-
The role of EZH1 and EZH2 in development and cancer.BMB Rep. 2022 Dec;55(12):595-601. doi: 10.5483/BMBRep.2022.55.12.174. BMB Rep. 2022. PMID: 36476271 Free PMC article. Review.
-
Structural basis for PRC2 engagement with chromatin.Curr Opin Struct Biol. 2021 Apr;67:135-144. doi: 10.1016/j.sbi.2020.10.017. Epub 2020 Nov 21. Curr Opin Struct Biol. 2021. PMID: 33232890 Review.
Cited by
-
Critical Roles of Polycomb Repressive Complexes in Transcription and Cancer.Int J Mol Sci. 2022 Aug 24;23(17):9574. doi: 10.3390/ijms23179574. Int J Mol Sci. 2022. PMID: 36076977 Free PMC article. Review.
-
PRC2 is high maintenance.Genes Dev. 2019 Aug 1;33(15-16):903-935. doi: 10.1101/gad.325050.119. Epub 2019 May 23. Genes Dev. 2019. PMID: 31123062 Free PMC article. Review.
-
Regulatory interactions between RNA and polycomb repressive complex 2.Mol Cell. 2014 Jul 17;55(2):171-85. doi: 10.1016/j.molcel.2014.05.009. Epub 2014 May 29. Mol Cell. 2014. PMID: 24882207 Free PMC article.
-
Single-molecule and in silico dissection of the interaction between Polycomb repressive complex 2 and chromatin.Proc Natl Acad Sci U S A. 2020 Dec 1;117(48):30465-30475. doi: 10.1073/pnas.2003395117. Epub 2020 Nov 18. Proc Natl Acad Sci U S A. 2020. PMID: 33208532 Free PMC article.
-
Protein lysine 43 methylation by EZH1 promotes AML1-ETO transcriptional repression in leukemia.Nat Commun. 2019 Nov 7;10(1):5051. doi: 10.1038/s41467-019-12960-6. Nat Commun. 2019. PMID: 31699991 Free PMC article.
References
-
- Bernstein BE, Mikkelsen TS, Xie X, Kamal M, Huebert DJ, Cuff J, Fry B, Meissner A, Wernig M, Plath K, et al. 2006. A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125: 315–326 - PubMed
-
- Brown JL, Mucci D, Whiteley M, Dirksen ML, Kassis JA 1998. The Drosophila Polycomb group gene pleiohomeotic encodes a DNA binding protein with homology to the transcription factor YY1. Mol Cell 1: 1057–1064 - PubMed
-
- Cao R, Zhang Y 2004. SUZ12 is required for both the histone methyltransferase activity and the silencing function of the EED–EZH2 complex. Mol Cell 15: 57–67 - PubMed
-
- Cao R, Wang L, Wang H, Xia L, Erdjument-Bromage H, Tempst P, Jones RS, Zhang Y 2002. Role of histone H3 lysine 27 methylation in Polycomb-group silencing. Science 298: 1039–1043 - PubMed
Publication types
MeSH terms
Substances
Associated data
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
