Genetic determinants and cellular constraints in noisy gene expression
- PMID: 24311680
- PMCID: PMC4045091
- DOI: 10.1126/science.1242975
Genetic determinants and cellular constraints in noisy gene expression
Abstract
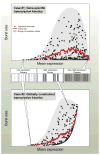
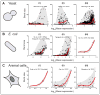
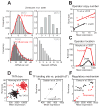
In individual cells, transcription is a random process obeying single-molecule kinetics. Often, it occurs in a bursty, intermittent manner. The frequency and size of these bursts affect the magnitude of temporal fluctuations in messenger RNA and protein content within a cell, creating variation or "noise" in gene expression. It is still unclear to what degree transcriptional kinetics are specific to each gene and determined by its promoter sequence. Alternative scenarios have been proposed, in which the kinetics of transcription are governed by cellular constraints and follow universal rules across the genome. Evidence from genome-wide noise studies and from systematic perturbations of promoter sequences suggest that both scenarios-namely gene-specific versus genome-wide regulation of transcription kinetics-may be present to different degrees in bacteria, yeast, and animal cells.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures
Similar articles
-
The route to transcription initiation determines the mode of transcriptional bursting in E. coli.Nat Commun. 2020 May 15;11(1):2422. doi: 10.1038/s41467-020-16367-6. Nat Commun. 2020. PMID: 32415118 Free PMC article.
-
Stationary moments, distribution conjugation and phenotypic regions in stochastic gene transcription.Math Biosci Eng. 2019 Jul 3;16(5):6134-6166. doi: 10.3934/mbe.2019307. Math Biosci Eng. 2019. PMID: 31499756
-
Cis-Regulatory Logic Produces Gene-Expression Noise Describing Phenotypic Heterogeneity in Bacteria.Front Genet. 2021 Sep 28;12:698910. doi: 10.3389/fgene.2021.698910. eCollection 2021. Front Genet. 2021. PMID: 34650591 Free PMC article. Review.
-
Non-equilibrium dynamics of stochastic gene regulation.J Biol Phys. 2015 Jan;41(1):49-58. doi: 10.1007/s10867-014-9365-9. Epub 2014 Oct 8. J Biol Phys. 2015. PMID: 25288134 Free PMC article.
-
Stochastic transcription initiation: Time dependent transcription rates.Biophys Chem. 2006 Apr 20;121(1):51-6. doi: 10.1016/j.bpc.2005.12.010. Epub 2006 Jan 27. Biophys Chem. 2006. PMID: 16442697 Review.
Cited by
-
RNA imaging. Spatially resolved, highly multiplexed RNA profiling in single cells.Science. 2015 Apr 24;348(6233):aaa6090. doi: 10.1126/science.aaa6090. Epub 2015 Apr 9. Science. 2015. PMID: 25858977 Free PMC article.
-
Collective polymerase dynamics emerge from DNA supercoiling during transcription.Biophys J. 2022 Nov 1;121(21):4153-4165. doi: 10.1016/j.bpj.2022.09.026. Epub 2022 Sep 27. Biophys J. 2022. PMID: 36171726 Free PMC article.
-
Unravelling the Roles of Bacterial Nanomachines Bistability in Pathogens' Life Cycle.Microorganisms. 2024 Sep 23;12(9):1930. doi: 10.3390/microorganisms12091930. Microorganisms. 2024. PMID: 39338604 Free PMC article. Review.
-
A stochastic model for post-transcriptional regulation of rare events in gene expression.Phys Biol. 2019 Apr 12;16(4):045003. doi: 10.1088/1478-3975/aafbef. Phys Biol. 2019. PMID: 30609418 Free PMC article.
-
Challenges in measuring and understanding biological noise.Nat Rev Genet. 2019 Sep;20(9):536-548. doi: 10.1038/s41576-019-0130-6. Nat Rev Genet. 2019. PMID: 31114032 Free PMC article. Review.
References
-
- Yunger S, Rosenfeld L, Garini Y, Shav-Tal Y. Single-allele analysis of transcription kinetics in living mammalian cells. Nat Methods. 2010;7:631–633. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

