The crosstalk between plant microRNA biogenesis factors and the spliceosome
- PMID: 24300047
- PMCID: PMC4091587
- DOI: 10.4161/psb.26955
The crosstalk between plant microRNA biogenesis factors and the spliceosome
Abstract
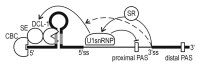
Many of the plant microRNA (miRNA) genes contain introns. The miRNA-containing hairpin loop structure is predominantly located within the first exon of such pri-miRNAs. We have shown that the downstream intron and its splicing are important for the regulation of the processing of these pri-miRNAs. The 5' splice site in MIR genes is essential in the process of miRNA biogenesis. We postulate that the presence of yet undefined interactions between U1 snRNP and the pri-miRNA processing machinery leads to an increase in the efficiency of miRNA biogenesis. The 5' splice site also decreases the accessibility of the polyadenylation machinery to the pri-miRNA polyA signal located within the same intron. It is likely that the spliceosome assembly controls the length and structure of MIR primary transcripts, and regulates mature miRNA levels. The emerging picture shows that introns, splicing, and/or alternative splicing have highly relevant roles in regulating the miRNA levels in very specific conditions that contribute to proper plant response to stress conditions.
Keywords: intron-containing pri-miRNAs; plant microRNA biogenesis; polyadenylation; splicing.
Figures
Similar articles
-
Posttranscriptional coordination of splicing and miRNA biogenesis in plants.Wiley Interdiscip Rev RNA. 2017 May;8(3). doi: 10.1002/wrna.1403. Epub 2016 Nov 9. Wiley Interdiscip Rev RNA. 2017. PMID: 27863087 Review.
-
Spliceosome disassembly factors ILP1 and NTR1 promote miRNA biogenesis in Arabidopsis thaliana.Nucleic Acids Res. 2019 Sep 5;47(15):7886-7900. doi: 10.1093/nar/gkz526. Nucleic Acids Res. 2019. PMID: 31216029 Free PMC article.
-
Alternative splicing regulates biogenesis of miRNAs located across exon-intron junctions.Mol Cell. 2013 Jun 27;50(6):869-81. doi: 10.1016/j.molcel.2013.05.007. Epub 2013 Jun 6. Mol Cell. 2013. PMID: 23747012
-
Primary microRNA transcript retention at sites of transcription leads to enhanced microRNA production.J Cell Biol. 2008 Jul 14;182(1):61-76. doi: 10.1083/jcb.200803111. J Cell Biol. 2008. PMID: 18625843 Free PMC article.
-
Cross talk between spliceosome and microprocessor defines the fate of pre-mRNA.Wiley Interdiscip Rev RNA. 2014 Sep-Oct;5(5):647-58. doi: 10.1002/wrna.1236. Epub 2014 Apr 30. Wiley Interdiscip Rev RNA. 2014. PMID: 24788135 Review.
Cited by
-
Short intron-derived ncRNAs.Nucleic Acids Res. 2017 May 5;45(8):4768-4781. doi: 10.1093/nar/gkw1341. Nucleic Acids Res. 2017. PMID: 28053119 Free PMC article.
-
The PROTEIN PHOSPHATASE4 Complex Promotes Transcription and Processing of Primary microRNAs in Arabidopsis.Plant Cell. 2019 Feb;31(2):486-501. doi: 10.1105/tpc.18.00556. Epub 2019 Jan 23. Plant Cell. 2019. PMID: 30674692 Free PMC article.
-
miR393 contributes to the embryogenic transition induced in vitro in Arabidopsis via the modification of the tissue sensitivity to auxin treatment.Planta. 2016 Jul;244(1):231-43. doi: 10.1007/s00425-016-2505-7. Epub 2016 Apr 4. Planta. 2016. PMID: 27040841 Free PMC article.
-
Cold-Dependent Expression and Alternative Splicing of Arabidopsis Long Non-coding RNAs.Front Plant Sci. 2019 Feb 28;10:235. doi: 10.3389/fpls.2019.00235. eCollection 2019. Front Plant Sci. 2019. PMID: 30891054 Free PMC article.
-
Core spliceosomal Sm proteins as constituents of cytoplasmic mRNPs in plants.Plant J. 2020 Aug;103(3):1155-1173. doi: 10.1111/tpj.14792. Epub 2020 May 28. Plant J. 2020. PMID: 32369637 Free PMC article.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
