The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids
- PMID: 24270517
- PMCID: PMC3896265
- DOI: 10.1038/ni.2767
The miR-126-VEGFR2 axis controls the innate response to pathogen-associated nucleic acids
Abstract
miR-126 is a microRNA expressed predominately by endothelial cells and controls angiogenesis. We found miR-126 was required for the innate response to pathogen-associated nucleic acids and that miR-126-deficient mice had greater susceptibility to infection with pseudotyped HIV. Profiling of miRNA indicated that miR-126 had high and specific expression by plasmacytoid dendritic cells (pDCs). Moreover, miR-126 controlled the survival and function of pDCs and regulated the expression of genes encoding molecules involved in the innate response, including Tlr7, Tlr9 and Nfkb1, as well as Kdr, which encodes the growth factor receptor VEGFR2. Deletion of Kdr in DCs resulted in reduced production of type I interferon, which supports the proposal of a role for VEGFR2 in miR-126 regulation of pDCs. Our studies identify the miR-126-VEGFR2 axis as an important regulator of the innate response that operates through multiscale control of pDCs.
Figures
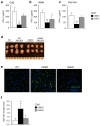
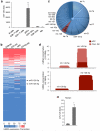


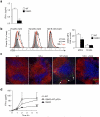
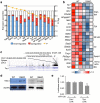
Comment in
-
Innate immunity: A surprise regulator of plasmacytoid DCs.Nat Rev Immunol. 2014 Jan;14(1):2-3. doi: 10.1038/nri3590. Epub 2013 Dec 13. Nat Rev Immunol. 2014. PMID: 24336100 No abstract available.
-
A new VEGF connection between two old neighbors.Nat Immunol. 2014 Jan;15(1):8-9. doi: 10.1038/ni.2786. Nat Immunol. 2014. PMID: 24352317 Free PMC article.
-
One microRNA controls both angiogenesis and TLR-mediated innate immunity to nucleic acids.Mol Ther. 2014 Feb;22(2):249-250. doi: 10.1038/mt.2013.299. Mol Ther. 2014. PMID: 24487566 Free PMC article. No abstract available.
-
miR-126, a new modulator of innate immunity.Cell Mol Immunol. 2014 May;11(3):215-7. doi: 10.1038/cmi.2014.5. Epub 2014 Feb 17. Cell Mol Immunol. 2014. PMID: 24531618 Free PMC article. No abstract available.
Similar articles
-
Bruton's tyrosine kinase regulates TLR9 but not TLR7 signaling in human plasmacytoid dendritic cells.Eur J Immunol. 2014 Apr;44(4):1130-6. doi: 10.1002/eji.201344030. Epub 2014 Jan 20. Eur J Immunol. 2014. PMID: 24375473
-
PACSIN1 regulates the TLR7/9-mediated type I interferon response in plasmacytoid dendritic cells.Eur J Immunol. 2012 Mar;42(3):573-9. doi: 10.1002/eji.201142045. Eur J Immunol. 2012. PMID: 22488361 Free PMC article.
-
Autoimmune skin inflammation is dependent on plasmacytoid dendritic cell activation by nucleic acids via TLR7 and TLR9.J Exp Med. 2010 Dec 20;207(13):2931-42. doi: 10.1084/jem.20101048. Epub 2010 Nov 29. J Exp Med. 2010. PMID: 21115693 Free PMC article.
-
Impaired Toll-like receptor 7 and 9 signaling: from chronic viral infections to cancer.Trends Immunol. 2010 Oct;31(10):391-7. doi: 10.1016/j.it.2010.07.004. Epub 2010 Sep 9. Trends Immunol. 2010. PMID: 20832362 Review.
-
Molecular mechanisms for plasmacytoid dendritic cell function and development.Vaccine. 2010 Nov 23;28(50):8046-7. doi: 10.1016/j.vaccine.2010.09.025. Epub 2010 Sep 17. Vaccine. 2010. PMID: 20849871 Review.
Cited by
-
Exosomes miR-126a released from MDSC induced by DOX treatment promotes lung metastasis.Oncogene. 2017 Feb 2;36(5):639-651. doi: 10.1038/onc.2016.229. Epub 2016 Jun 27. Oncogene. 2017. PMID: 27345402 Free PMC article.
-
MicroRNA-155 Mediates Augmented CD40 Expression in Bone Marrow Derived Plasmacytoid Dendritic Cells in Symptomatic Lupus-Prone NZB/W F1 Mice.Int J Mol Sci. 2016 Aug 6;17(8):1282. doi: 10.3390/ijms17081282. Int J Mol Sci. 2016. PMID: 27509492 Free PMC article.
-
MicroRNAs as Important Players in Host-Adenovirus Interactions.Front Microbiol. 2017 Jul 17;8:1324. doi: 10.3389/fmicb.2017.01324. eCollection 2017. Front Microbiol. 2017. PMID: 28769895 Free PMC article. Review.
-
Microvesicles Derived from Indoxyl Sulfate Treated Endothelial Cells Induce Endothelial Progenitor Cells Dysfunction.Front Physiol. 2017 Sep 8;8:666. doi: 10.3389/fphys.2017.00666. eCollection 2017. Front Physiol. 2017. PMID: 28951723 Free PMC article.
-
MicroRNA-126 deficiency enhanced the activation and function of CD4+ T cells by elevating IRS-1 pathway.Clin Exp Immunol. 2018 Feb;191(2):166-179. doi: 10.1111/cei.13067. Epub 2017 Oct 30. Clin Exp Immunol. 2018. PMID: 28987000 Free PMC article.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

