Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism
- PMID: 24267887
- PMCID: PMC3934107
- DOI: 10.1016/j.cell.2013.10.031
Integrative functional genomic analyses implicate specific molecular pathways and circuits in autism
Abstract
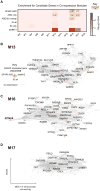
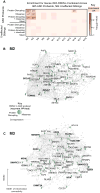
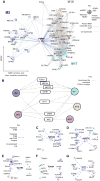
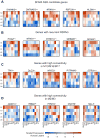
Genetic studies have identified dozens of autism spectrum disorder (ASD) susceptibility genes, raising two critical questions: (1) do these genetic loci converge on specific biological processes, and (2) where does the phenotypic specificity of ASD arise, given its genetic overlap with intellectual disability (ID)? To address this, we mapped ASD and ID risk genes onto coexpression networks representing developmental trajectories and transcriptional profiles representing fetal and adult cortical laminae. ASD genes tightly coalesce in modules that implicate distinct biological functions during human cortical development, including early transcriptional regulation and synaptic development. Bioinformatic analyses suggest that translational regulation by FMRP and transcriptional coregulation by common transcription factors connect these processes. At a circuit level, ASD genes are enriched in superficial cortical layers and glutamatergic projection neurons. Furthermore, we show that the patterns of ASD and ID risk genes are distinct, providing a biological framework for further investigating the pathophysiology of ASD.
Copyright © 2013 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Network- and attribute-based classifiers can prioritize genes and pathways for autism spectrum disorders and intellectual disability.Am J Med Genet C Semin Med Genet. 2012 May 15;160C(2):130-42. doi: 10.1002/ajmg.c.31330. Epub 2012 Apr 12. Am J Med Genet C Semin Med Genet. 2012. PMID: 22499558 Free PMC article.
-
Coexpression networks implicate human midfetal deep cortical projection neurons in the pathogenesis of autism.Cell. 2013 Nov 21;155(5):997-1007. doi: 10.1016/j.cell.2013.10.020. Cell. 2013. PMID: 24267886 Free PMC article.
-
CHD8 regulates neurodevelopmental pathways associated with autism spectrum disorder in neural progenitors.Proc Natl Acad Sci U S A. 2014 Oct 21;111(42):E4468-77. doi: 10.1073/pnas.1405266111. Epub 2014 Oct 7. Proc Natl Acad Sci U S A. 2014. PMID: 25294932 Free PMC article.
-
Glutamatergic candidate genes in autism spectrum disorder: an overview.J Neural Transm (Vienna). 2014 Sep;121(9):1081-106. doi: 10.1007/s00702-014-1161-y. Epub 2014 Feb 4. J Neural Transm (Vienna). 2014. PMID: 24493018 Review.
-
Intellectual disability and autism spectrum disorders: causal genes and molecular mechanisms.Neurosci Biobehav Rev. 2014 Oct;46 Pt 2:161-74. doi: 10.1016/j.neubiorev.2014.02.015. Epub 2014 Apr 4. Neurosci Biobehav Rev. 2014. PMID: 24709068 Free PMC article. Review.
Cited by
-
Immune mediators in the brain and peripheral tissues in autism spectrum disorder.Nat Rev Neurosci. 2015 Aug;16(8):469-86. doi: 10.1038/nrn3978. Nat Rev Neurosci. 2015. PMID: 26189694 Free PMC article. Review.
-
A Cre-dependent massively parallel reporter assay allows for cell-type specific assessment of the functional effects of non-coding elements in vivo.Commun Biol. 2023 Nov 13;6(1):1151. doi: 10.1038/s42003-023-05483-w. Commun Biol. 2023. PMID: 37953348 Free PMC article.
-
Pathogenic variants associated with speech/cognitive delay and seizures affect genes with expression biases in excitatory neurons and microglia in developing human cortex.bioRxiv [Preprint]. 2024 Jul 2:2024.07.01.601597. doi: 10.1101/2024.07.01.601597. bioRxiv. 2024. PMID: 39005386 Free PMC article. Preprint.
-
GABA/Glutamate synaptic pathways targeted by integrative genomic and electrophysiological explorations distinguish autism from intellectual disability.Mol Psychiatry. 2016 Mar;21(3):411-8. doi: 10.1038/mp.2015.75. Epub 2015 Jun 9. Mol Psychiatry. 2016. PMID: 26055424
-
Systems biology and gene networks in neurodevelopmental and neurodegenerative disorders.Nat Rev Genet. 2015 Aug;16(8):441-58. doi: 10.1038/nrg3934. Epub 2015 Jul 7. Nat Rev Genet. 2015. PMID: 26149713 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
- R01 MH094714/MH/NIMH NIH HHS/United States
- T32 MH073526/MH/NIMH NIH HHS/United States
- T32MH073526/MH/NIMH NIH HHS/United States
- 9R01MH100027/MH/NIMH NIH HHS/United States
- P30 NS062691/NS/NINDS NIH HHS/United States
- 5R37MH060233/MH/NIMH NIH HHS/United States
- RC2 MH089921/MH/NIMH NIH HHS/United States
- R01 MH060233/MH/NIMH NIH HHS/United States
- 5R01MH094714/MH/NIMH NIH HHS/United States
- F30 MH099886/MH/NIMH NIH HHS/United States
- F30MH099886/MH/NIMH NIH HHS/United States
- R01 MH100027/MH/NIMH NIH HHS/United States
- P30NS062691/NS/NINDS NIH HHS/United States
- R37 MH060233/MH/NIMH NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources

