Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology
- PMID: 24264436
- PMCID: PMC4059406
- DOI: 10.1038/ncomms3823
Fate tracing reveals hepatic stellate cells as dominant contributors to liver fibrosis independent of its aetiology
Abstract
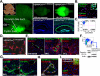
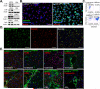
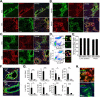
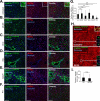
Although organ fibrosis causes significant morbidity and mortality in chronic diseases, the lack of detailed knowledge about specific cellular contributors mediating fibrogenesis hampers the design of effective antifibrotic therapies. Different cellular sources, including tissue-resident and bone marrow-derived fibroblasts, pericytes and epithelial cells, have been suggested to give rise to myofibroblasts, but their relative contributions remain controversial, with profound differences between organs and different diseases. Here we employ a novel Cre-transgenic mouse that marks 99% of hepatic stellate cells (HSCs), a liver-specific pericyte population, to demonstrate that HSCs give rise to 82-96% of myofibroblasts in models of toxic, cholestatic and fatty liver disease. Moreover, we exclude that HSCs function as facultative epithelial progenitor cells in the injured liver. On the basis these findings, HSCs should be considered the primary cellular target for antifibrotic therapies across all types of liver disease.
Figures


Comment in
-
Liver: hepatic stellate cells hold the key to liver fibrosis.Nat Rev Gastroenterol Hepatol. 2014 Feb;11(2):74. doi: 10.1038/nrgastro.2013.244. Epub 2013 Dec 10. Nat Rev Gastroenterol Hepatol. 2014. PMID: 24322897 No abstract available.
-
Novel fate-tracing strategies show that hepatic stellate cells mediate fibrosis in vivo.Gastroenterology. 2014 Jun;146(7):1823-5. doi: 10.1053/j.gastro.2014.04.010. Epub 2014 Apr 21. Gastroenterology. 2014. PMID: 24768331 Free PMC article. No abstract available.
Similar articles
-
Origin of myofibroblasts in the fibrotic liver in mice.Proc Natl Acad Sci U S A. 2014 Aug 12;111(32):E3297-305. doi: 10.1073/pnas.1400062111. Epub 2014 Jul 29. Proc Natl Acad Sci U S A. 2014. PMID: 25074909 Free PMC article.
-
Hedgehog-YAP Signaling Pathway Regulates Glutaminolysis to Control Activation of Hepatic Stellate Cells.Gastroenterology. 2018 Apr;154(5):1465-1479.e13. doi: 10.1053/j.gastro.2017.12.022. Epub 2018 Jan 3. Gastroenterology. 2018. PMID: 29305935 Free PMC article.
-
Liver fibrosis.Turk J Gastroenterol. 2018 Jan;29(1):14-21. doi: 10.5152/tjg.2018.17330. Turk J Gastroenterol. 2018. PMID: 29391303 Free PMC article. Review.
-
EphB2 receptor tyrosine kinase promotes hepatic fibrogenesis in mice via activation of hepatic stellate cells.Sci Rep. 2018 Feb 7;8(1):2532. doi: 10.1038/s41598-018-20926-9. Sci Rep. 2018. PMID: 29416088 Free PMC article.
-
Origin and function of myofibroblasts in the liver.Semin Liver Dis. 2015 May;35(2):97-106. doi: 10.1055/s-0035-1550061. Epub 2015 May 14. Semin Liver Dis. 2015. PMID: 25974896 Review.
Cited by
-
p62/SQSTM1 by Binding to Vitamin D Receptor Inhibits Hepatic Stellate Cell Activity, Fibrosis, and Liver Cancer.Cancer Cell. 2016 Oct 10;30(4):595-609. doi: 10.1016/j.ccell.2016.09.004. Cancer Cell. 2016. PMID: 27728806 Free PMC article.
-
A new fluorescence-based optical imaging method to non-invasively monitor hepatic myofibroblasts in vivo.J Hepatol. 2016 Jul;65(1):75-83. doi: 10.1016/j.jhep.2016.03.021. Epub 2016 Apr 9. J Hepatol. 2016. PMID: 27067455 Free PMC article.
-
Pentoxifylline inhibits liver fibrosis via hedgehog signaling pathway.J Huazhong Univ Sci Technolog Med Sci. 2016 Jun;36(3):372-376. doi: 10.1007/s11596-016-1594-7. Epub 2016 Jul 5. J Huazhong Univ Sci Technolog Med Sci. 2016. PMID: 27376806
-
In vitro reversion of activated primary human hepatic stellate cells.Fibrogenesis Tissue Repair. 2015 Aug 6;8:14. doi: 10.1186/s13069-015-0031-z. eCollection 2015. Fibrogenesis Tissue Repair. 2015. PMID: 26251672 Free PMC article.
-
Obesity and cancer-extracellular matrix, angiogenesis, and adrenergic signaling as unusual suspects linking the two diseases.Cancer Metastasis Rev. 2022 Sep;41(3):517-547. doi: 10.1007/s10555-022-10058-y. Epub 2022 Sep 8. Cancer Metastasis Rev. 2022. PMID: 36074318 Free PMC article. Review.
References
-
- Gabbiani G. The myofibroblast in wound healing and fibrocontractive diseases. J Pathol. 2003;200:500–503. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

