Transformation by Hras(G12V) is consistently associated with mutant allele copy gains and is reversed by farnesyl transferase inhibition
- PMID: 24240680
- PMCID: PMC4025988
- DOI: 10.1038/onc.2013.489
Transformation by Hras(G12V) is consistently associated with mutant allele copy gains and is reversed by farnesyl transferase inhibition
Abstract
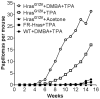
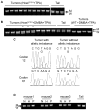
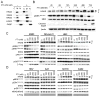
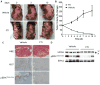
RAS-driven malignancies remain a major therapeutic challenge. The two-stage 7,12-dimethylbenz(a)anthracene (DMBA)/12-o-tetradecanoylphorbol-13-acetate (TPA) model of mouse skin carcinogenesis has been used to study mechanisms of epithelial tumor development by oncogenic Hras. We used mice with an Hras(G12V) knock-in allele to elucidate the early events after Hras activation, and to evaluate the therapeutic effectiveness of farnesyltransferase inhibition (FTI). Treatment of Caggs-Cre/FR-Hras(G12V) mice with TPA alone was sufficient to trigger papilloma development with a shorter latency and an ∼10-fold greater tumor burden than DMBA/TPA-treated WT-controls. Hras(G12V) allele copy number was increased in all papillomas induced by TPA. DMBA/TPA treatment of Hras(G12V) knock-in mice induced an even greater incidence of papillomas, which either harbored Hras(G12V) amplification or developed an Hras(Q61L) mutation in the second allele. Laser-capture microdissection of normal skin, hyperplastic skin and papillomas showed that amplification occurred only at the papilloma stage. HRAS-mutant allelic imbalance was also observed in human cancer cell lines, consistent with a requirement for augmented oncogenic HRAS signaling for tumor development. The FTI SCH66336 blocks HRAS farnesylation and delocalizes it from the plasma membrane. NRAS and KRAS are not affected as they are alternatively prenylated. When tested in lines harboring HRAS, NRAS or KRAS mutations, SCH66336 delocalized, inhibited signaling and preferentially inhibited growth only of HRAS-mutant lines. Treatment with SCH66336 also induced near-complete regression of papillomas of TPA-treated Hras(G12V) knock-in mice. These data suggest that farnesyl transferase inhibitors should be reevaluated as targeted agents for human HRAS-driven cancers, such as those of bladder, thyroid and other epithelial lineages.
Figures

Similar articles
-
Genetic alterations cooperate with v-Ha-ras to accelerate multistage carcinogenesis in TG.AC transgenic mouse skin.Cancer Res. 1995 Jul 15;55(14):3171-8. Cancer Res. 1995. PMID: 7606738
-
A farnesyl transferase inhibitor suppresses TPA-mediated skin tumor development without altering hyperplasia in the ras transgenic Tg.AC mouse.Mol Carcinog. 2000 Jan;27(1):24-33. doi: 10.1002/(sici)1098-2744(200001)27:1<24::aid-mc5>3.0.co;2-m. Mol Carcinog. 2000. PMID: 10642434
-
Evidence that mirex promotes a unique population of epidermal cells that cannot be distinguished by their mutant Ha-ras genotype.Mol Carcinog. 1997 Sep;20(1):115-24. Mol Carcinog. 1997. PMID: 9328442
-
Non-melanoma skin cancer in mouse and man.Arch Toxicol. 2013 May;87(5):783-98. doi: 10.1007/s00204-012-0998-9. Epub 2012 Dec 25. Arch Toxicol. 2013. PMID: 23266722 Review.
-
Skin cancer chemoprevention by α-santalol.Front Biosci (Schol Ed). 2011 Jan 1;3(2):777-87. doi: 10.2741/s186. Front Biosci (Schol Ed). 2011. PMID: 21196411 Review.
Cited by
-
Targeting the ERK Signaling Pathway in Melanoma.Int J Mol Sci. 2019 Mar 25;20(6):1483. doi: 10.3390/ijms20061483. Int J Mol Sci. 2019. PMID: 30934534 Free PMC article. Review.
-
Beyond EGFR Targeting in SCCHN: Angiogenesis, PI3K, and Other Molecular Targets.Front Oncol. 2019 Feb 13;9:74. doi: 10.3389/fonc.2019.00074. eCollection 2019. Front Oncol. 2019. PMID: 30815390 Free PMC article. Review.
-
Transposon mutagenesis identifies chromatin modifiers cooperating with Ras in thyroid tumorigenesis and detects ATXN7 as a cancer gene.Proc Natl Acad Sci U S A. 2017 Jun 20;114(25):E4951-E4960. doi: 10.1073/pnas.1702723114. Epub 2017 Jun 5. Proc Natl Acad Sci U S A. 2017. PMID: 28584132 Free PMC article.
-
The Role of Wild-Type RAS in Oncogenic RAS Transformation.Genes (Basel). 2021 Apr 28;12(5):662. doi: 10.3390/genes12050662. Genes (Basel). 2021. PMID: 33924994 Free PMC article. Review.
-
New tricks for human farnesyltransferase inhibitor: cancer and beyond.Medchemcomm. 2017 Feb 16;8(5):841-854. doi: 10.1039/c7md00030h. eCollection 2017 May 1. Medchemcomm. 2017. PMID: 30108801 Free PMC article. Review.
References
-
- Schubbert S, Shannon K, Bollag G. Hyperactive Ras in developmental disorders and cancer. Nat Rev Cancer. 2007;7:295–308. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

