TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein
- PMID: 24227843
- PMCID: PMC3911672
- DOI: 10.1128/JVI.02202-13
TMPRSS2 and ADAM17 cleave ACE2 differentially and only proteolysis by TMPRSS2 augments entry driven by the severe acute respiratory syndrome coronavirus spike protein
Abstract
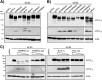
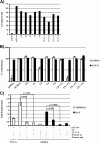
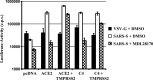
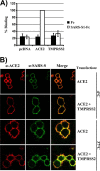
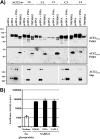
The type II transmembrane serine proteases TMPRSS2 and HAT can cleave and activate the spike protein (S) of the severe acute respiratory syndrome coronavirus (SARS-CoV) for membrane fusion. In addition, these proteases cleave the viral receptor, the carboxypeptidase angiotensin-converting enzyme 2 (ACE2), and it was proposed that ACE2 cleavage augments viral infectivity. However, no mechanistic insights into this process were obtained and the relevance of ACE2 cleavage for SARS-CoV S protein (SARS-S) activation has not been determined. Here, we show that arginine and lysine residues within ACE2 amino acids 697 to 716 are essential for cleavage by TMPRSS2 and HAT and that ACE2 processing is required for augmentation of SARS-S-driven entry by these proteases. In contrast, ACE2 cleavage was dispensable for activation of the viral S protein. Expression of TMPRSS2 increased cellular uptake of soluble SARS-S, suggesting that protease-dependent augmentation of viral entry might be due to increased uptake of virions into target cells. Finally, TMPRSS2 was found to compete with the metalloprotease ADAM17 for ACE2 processing, but only cleavage by TMPRSS2 resulted in augmented SARS-S-driven entry. Collectively, our results in conjunction with those of previous studies indicate that TMPRSS2 and potentially related proteases promote SARS-CoV entry by two separate mechanisms: ACE2 cleavage, which might promote viral uptake, and SARS-S cleavage, which activates the S protein for membrane fusion. These observations have interesting implications for the development of novel therapeutics. In addition, they should spur efforts to determine whether receptor cleavage promotes entry of other coronaviruses, which use peptidases as entry receptors.
Figures



Similar articles
-
SARS-CoV-2 and SARS-CoV Spike-Mediated Cell-Cell Fusion Differ in Their Requirements for Receptor Expression and Proteolytic Activation.J Virol. 2021 Apr 12;95(9):e00002-21. doi: 10.1128/JVI.00002-21. Print 2021 Apr 12. J Virol. 2021. PMID: 33608407 Free PMC article.
-
SARS-CoV-2 Cell Entry Depends on ACE2 and TMPRSS2 and Is Blocked by a Clinically Proven Protease Inhibitor.Cell. 2020 Apr 16;181(2):271-280.e8. doi: 10.1016/j.cell.2020.02.052. Epub 2020 Mar 5. Cell. 2020. PMID: 32142651 Free PMC article.
-
Distinctive Roles of Furin and TMPRSS2 in SARS-CoV-2 Infectivity.J Virol. 2022 Apr 27;96(8):e0012822. doi: 10.1128/jvi.00128-22. Epub 2022 Mar 28. J Virol. 2022. PMID: 35343766 Free PMC article.
-
Proteolytic activation of the SARS-coronavirus spike protein: cutting enzymes at the cutting edge of antiviral research.Antiviral Res. 2013 Dec;100(3):605-14. doi: 10.1016/j.antiviral.2013.09.028. Epub 2013 Oct 8. Antiviral Res. 2013. PMID: 24121034 Free PMC article. Review.
-
Contributions of human ACE2 and TMPRSS2 in determining host-pathogen interaction of COVID-19.J Genet. 2021;100(1):12. doi: 10.1007/s12041-021-01262-w. J Genet. 2021. PMID: 33707363 Free PMC article. Review.
Cited by
-
Coronavirus and influenza virus proteolytic priming takes place in tetraspanin-enriched membrane microdomains.J Virol. 2015 Jun;89(11):6093-104. doi: 10.1128/JVI.00543-15. Epub 2015 Apr 1. J Virol. 2015. PMID: 25833045 Free PMC article.
-
ACEIs and ARBs and Their Correlation with COVID-19: A Review.Infect Drug Resist. 2020 Sep 16;13:3217-3224. doi: 10.2147/IDR.S264882. eCollection 2020. Infect Drug Resist. 2020. PMID: 32982336 Free PMC article. Review.
-
Impact of Comorbidities on SARS-CoV-2 Viral Entry-Related Genes.J Pers Med. 2020 Sep 25;10(4):146. doi: 10.3390/jpm10040146. J Pers Med. 2020. PMID: 32992731 Free PMC article.
-
Nucleoside Analogs and Nucleoside Precursors as Drugs in the Fight against SARS-CoV-2 and Other Coronaviruses.Molecules. 2021 Feb 13;26(4):986. doi: 10.3390/molecules26040986. Molecules. 2021. PMID: 33668428 Free PMC article. Review.
-
Should ACE2 be given a chance in COVID-19 therapeutics: A semi-systematic review of strategies enhancing ACE2.Eur J Pharmacol. 2020 Nov 15;887:173545. doi: 10.1016/j.ejphar.2020.173545. Epub 2020 Sep 11. Eur J Pharmacol. 2020. PMID: 32926917 Free PMC article. Review.
References
-
- Holmes KV. 2001. Coronaviruses, p 1187–1203 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed). 2001. Fields virology, 4th ed. Lippincott Williams & Wilkins, Philadelphia, PA
-
- Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, Wong BH, Poon RW, Cai JJ, Luk WK, Poon LL, Wong SS, Guan Y, Peiris JS, Yuen KY. 2005. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J. Virol. 79:884–895. 10.1128/JVI.79.2.884-895.2005 - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

