Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes
- PMID: 24213554
- PMCID: PMC3962538
- DOI: 10.4161/epi.27021
Epigenomic elements enriched in the promoters of autoimmunity susceptibility genes
Abstract
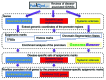
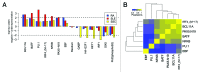
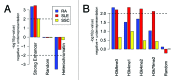
Genome-wide association studies have identified a number of autoimmune disease-susceptibility genes. Whether or not these loci share any regulatory or functional elements, however, is an open question. Finding such common regulators is of considerable research interest in order to define systemic therapeutic targets. The growing amount of experimental genomic annotations, particularly those from the ENCODE project, provide a wealth of opportunities to search for such commonalities. We hypothesized that regulatory commonalities might not only delineate a regulatory landscape predisposing to autoimmune diseases, but also define functional elements distinguishing specific diseases. We further investigated if, and how, disease-specific epigenomic elements can identify novel genes yet to be associated with the diseases. We evaluated transcription factors, histone modifications, and chromatin state data obtained from the ENCODE project for statistically significant over- or under-representation in the promoters of genes associated with Systemic Lupus Erythematosus (SLE), Rheumatoid Arthritis (RA), and Systemic Sclerosis (SSc). We identified BATF, BCL11A, IRF4, NFkB, PAX5, and PU.1 as transcription factors over-represented in SLE- and RA-susceptibility gene promoters. H3K4me1 and H3K4me2 epigenomic marks were associated with SLE susceptibility genes, and H3K9me3 was common to both SLE and RA. In contrast to a transcriptionally active signature in SLE and RA, SSc-susceptibility genes were depleted in activating epigenomic elements. Using epigenomic elements enriched in SLE and RA, we identified additional immune and B cell signaling-related genes with the same elements in their promoters. Our analysis suggests common and disease-specific epigenomic elements that may define novel therapeutic targets for controlling aberrant activation of autoimmune susceptibility genes.
Keywords: ENCODE; GenomeRunner; autoimmunity; epigenetics; genome.
Figures
Similar articles
-
The Pathogenic Role of Dysregulated Epigenetic Modifications in Autoimmune Diseases.Front Immunol. 2019 Sep 27;10:2305. doi: 10.3389/fimmu.2019.02305. eCollection 2019. Front Immunol. 2019. PMID: 31611879 Free PMC article. Review.
-
Epigenetic regulation of immune cells in systemic lupus erythematosus: insight from chromatin accessibility.Expert Opin Ther Targets. 2024 Jul;28(7):637-649. doi: 10.1080/14728222.2024.2375372. Epub 2024 Jul 2. Expert Opin Ther Targets. 2024. PMID: 38943564 Review.
-
A systemic sclerosis and systemic lupus erythematosus pan-meta-GWAS reveals new shared susceptibility loci.Hum Mol Genet. 2013 Oct 1;22(19):4021-9. doi: 10.1093/hmg/ddt248. Epub 2013 Jun 4. Hum Mol Genet. 2013. PMID: 23740937 Free PMC article.
-
Germline genetic patterns underlying familial rheumatoid arthritis, systemic lupus erythematosus and primary Sjögren's syndrome highlight T cell-initiated autoimmunity.Ann Rheum Dis. 2020 Feb;79(2):268-275. doi: 10.1136/annrheumdis-2019-215533. Epub 2019 Dec 17. Ann Rheum Dis. 2020. PMID: 31848144
-
Cross-Disease Innate Gene Signature: Emerging Diversity and Abundance in RA Comparing to SLE and SSc.J Immunol Res. 2019 Jul 16;2019:3575803. doi: 10.1155/2019/3575803. eCollection 2019. J Immunol Res. 2019. PMID: 31396542 Free PMC article.
Cited by
-
Integrating Epigenomic Elements and GWASs Identifies BDNF Gene Affecting Bone Mineral Density and Osteoporotic Fracture Risk.Sci Rep. 2016 Jul 28;6:30558. doi: 10.1038/srep30558. Sci Rep. 2016. PMID: 27465306 Free PMC article.
-
Multiple DNA-binding modes for the ETS family transcription factor PU.1.J Biol Chem. 2017 Sep 29;292(39):16044-16054. doi: 10.1074/jbc.M117.798207. Epub 2017 Aug 8. J Biol Chem. 2017. PMID: 28790174 Free PMC article.
-
Transcription Factor Activity Inference in Systemic Lupus Erythematosus.Life (Basel). 2021 Apr 1;11(4):299. doi: 10.3390/life11040299. Life (Basel). 2021. PMID: 33915751 Free PMC article.
-
Polycomb repressive complex 2 epigenomic signature defines age-associated hypermethylation and gene expression changes.Epigenetics. 2015;10(6):484-95. doi: 10.1080/15592294.2015.1040619. Epub 2015 Apr 16. Epigenetics. 2015. PMID: 25880792 Free PMC article.
-
The transmembrane adapter SCIMP recruits tyrosine kinase Syk to phosphorylate Toll-like receptors to mediate selective inflammatory outputs.J Biol Chem. 2022 May;298(5):101857. doi: 10.1016/j.jbc.2022.101857. Epub 2022 Mar 22. J Biol Chem. 2022. PMID: 35337798 Free PMC article.
References
-
- Rossin EJ, Lage K, Raychaudhuri S, Xavier RJ, Tatar D, Benita Y, Cotsapas C, Daly MJ, International Inflammatory Bowel Disease Genetics Constortium Proteins encoded in genomic regions associated with immune-mediated disease physically interact and suggest underlying biology. PLoS Genet. 2011;7:e1001273. doi: 10.1371/journal.pgen.1001273. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
