Persistently adenovirus-infected lymphoid cells express microRNAs derived from the viral VAI and especially VAII RNA
- PMID: 24210108
- PMCID: PMC3825519
- DOI: 10.1016/j.virol.2013.08.024
Persistently adenovirus-infected lymphoid cells express microRNAs derived from the viral VAI and especially VAII RNA
Abstract
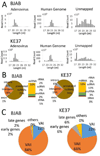
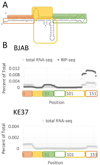
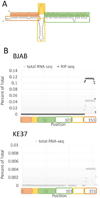
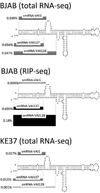
Human adenovirus can establish latent infections in lymphoid tissues in vivo and persistent, infections in cultured lymphoid cell lines. During lytic infection, adenovirus expresses microRNAs (miRNAs) derived from the viral non-coding RNAs VAI and, especially, VAII. Here, we demonstrate that persistently adenovirus-infected human BJAB cells also produce adenovirus-derived miRNAs primarily derived from the viral VAII RNA, which contributes ~2.7% of all RNA-induced silencing complex (RISC)-associated RNAs. However, our data indicate that the 5' end of the predominant VAII-derived viral RNA, and hence its seed sequence, differs from what has been previously reported. Our data demonstrate that adenovirus expresses viral miRNAs in chronically infected lymphoid cells and raise the possibility that these may contribute to the maintenance of the latently adenovirus-infected lymphoid cells previously observed in mucosal-associated lymphoid tissues in vivo.
Keywords: Adenovirus; MicroRNAs; Persistent infection; RNA interference.
© 2013 Elsevier Inc. All rights reserved.
Figures



Similar articles
-
Identification of RISC-associated adenoviral microRNAs, a subset of their direct targets, and global changes in the targetome upon lytic adenovirus 5 infection.J Virol. 2015 Feb;89(3):1608-27. doi: 10.1128/JVI.02336-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410853 Free PMC article.
-
Adenovirus mutants with DNA sequence perturbations in the intragenic promoter of VAI RNA gene allow the enhanced transcription of VAII RNA gene in HeLa cells.Nucleic Acids Res. 1984 Oct 11;12(19):7377-88. doi: 10.1093/nar/12.19.7377. Nucleic Acids Res. 1984. PMID: 6493978 Free PMC article.
-
Construction and analysis of additional adenovirus substitution mutants confirm the complementation of VAI RNA function by two small RNAs encoded by Epstein-Barr virus.J Virol. 1985 Dec;56(3):750-6. doi: 10.1128/JVI.56.3.750-756.1985. J Virol. 1985. PMID: 2999431 Free PMC article.
-
Adenovirus and miRNAs.Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):660-7. doi: 10.1016/j.bbagrm.2011.05.004. Epub 2011 May 17. Biochim Biophys Acta. 2011. PMID: 21621026 Free PMC article. Review.
-
Roles and regulation of microRNAs in cytomegalovirus infection.Biochim Biophys Acta. 2011 Nov-Dec;1809(11-12):613-22. doi: 10.1016/j.bbagrm.2011.04.002. Epub 2011 Apr 21. Biochim Biophys Acta. 2011. PMID: 21545855 Review.
Cited by
-
Characterization of Viral miRNAs during Adenovirus 14 Infection and Their Differential Expression in the Emergent Strain Adenovirus 14p1.Viruses. 2022 Apr 26;14(5):898. doi: 10.3390/v14050898. Viruses. 2022. PMID: 35632641 Free PMC article.
-
NAD-linked mechanisms of gene de-repression and a novel role for CtBP in persistent adenovirus infection of lymphocytes.Virol J. 2019 Dec 21;16(1):161. doi: 10.1186/s12985-019-1265-y. Virol J. 2019. PMID: 31864392 Free PMC article.
-
Simultaneous Single-Cell In Situ Analysis of Human Adenovirus Type 5 DNA and mRNA Expression Patterns in Lytic and Persistent Infection.J Virol. 2017 May 12;91(11):e00166-17. doi: 10.1128/JVI.00166-17. Print 2017 Jun 1. J Virol. 2017. PMID: 28298601 Free PMC article.
-
Synthesis, Structure, and Function of Human Adenovirus Small Non-Coding RNAs.Viruses. 2020 Oct 19;12(10):1182. doi: 10.3390/v12101182. Viruses. 2020. PMID: 33086737 Free PMC article. Review.
-
Epigenetics and the dynamics of chromatin during adenovirus infections.FEBS Lett. 2019 Dec;593(24):3551-3570. doi: 10.1002/1873-3468.13697. Epub 2019 Dec 15. FEBS Lett. 2019. PMID: 31769503 Free PMC article. Review.
References
-
- Avila MM, Carballal G, Rovaletti H, Ebekian B, Cusminsky M, Weissenbacher M. Viral etiology in acute lower respiratory infections in children from a closed community. Am Rev Respir Dis. 1989;140:634–637. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

