CanDrA: cancer-specific driver missense mutation annotation with optimized features
- PMID: 24205039
- PMCID: PMC3813554
- DOI: 10.1371/journal.pone.0077945
CanDrA: cancer-specific driver missense mutation annotation with optimized features
Abstract
Driver mutations are somatic mutations that provide growth advantage to tumor cells, while passenger mutations are those not functionally related to oncogenesis. Distinguishing drivers from passengers is challenging because drivers occur much less frequently than passengers, they tend to have low prevalence, their functions are multifactorial and not intuitively obvious. Missense mutations are excellent candidates as drivers, as they occur more frequently and are potentially easier to identify than other types of mutations. Although several methods have been developed for predicting the functional impact of missense mutations, only a few have been specifically designed for identifying driver mutations. As more mutations are being discovered, more accurate predictive models can be developed using machine learning approaches that systematically characterize the commonality and peculiarity of missense mutations under the background of specific cancer types. Here, we present a cancer driver annotation (CanDrA) tool that predicts missense driver mutations based on a set of 95 structural and evolutionary features computed by over 10 functional prediction algorithms such as CHASM, SIFT, and MutationAssessor. Through feature optimization and supervised training, CanDrA outperforms existing tools in analyzing the glioblastoma multiforme and ovarian carcinoma data sets in The Cancer Genome Atlas and the Cancer Cell Line Encyclopedia project.
Conflict of interest statement
Figures


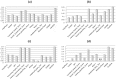
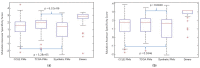
Similar articles
-
Computational Approaches to Prioritize Cancer Driver Missense Mutations.Int J Mol Sci. 2018 Jul 20;19(7):2113. doi: 10.3390/ijms19072113. Int J Mol Sci. 2018. PMID: 30037003 Free PMC article. Review.
-
Cancer-specific high-throughput annotation of somatic mutations: computational prediction of driver missense mutations.Cancer Res. 2009 Aug 15;69(16):6660-7. doi: 10.1158/0008-5472.CAN-09-1133. Epub 2009 Aug 4. Cancer Res. 2009. PMID: 19654296 Free PMC article.
-
Assessment of computational methods for predicting the effects of missense mutations in human cancers.BMC Genomics. 2013;14 Suppl 3(Suppl 3):S7. doi: 10.1186/1471-2164-14-S3-S7. Epub 2013 May 28. BMC Genomics. 2013. PMID: 23819521 Free PMC article.
-
Distinguishing between driver and passenger mutations in individual cancer genomes by network enrichment analysis.BMC Bioinformatics. 2014 Sep 19;15(1):308. doi: 10.1186/1471-2105-15-308. BMC Bioinformatics. 2014. PMID: 25236784 Free PMC article.
-
Identifying driver mutations from sequencing data of heterogeneous tumors in the era of personalized genome sequencing.Brief Bioinform. 2014 Mar;15(2):244-55. doi: 10.1093/bib/bbt042. Epub 2013 Jul 1. Brief Bioinform. 2014. PMID: 23818492 Review.
Cited by
-
A decision support framework for genomically informed investigational cancer therapy.J Natl Cancer Inst. 2015 Apr 11;107(7):djv098. doi: 10.1093/jnci/djv098. Print 2015 Jul. J Natl Cancer Inst. 2015. PMID: 25863335 Free PMC article. Review.
-
CancerVar: An artificial intelligence-empowered platform for clinical interpretation of somatic mutations in cancer.Sci Adv. 2022 May 6;8(18):eabj1624. doi: 10.1126/sciadv.abj1624. Epub 2022 May 6. Sci Adv. 2022. PMID: 35544644 Free PMC article.
-
Implementation of biomarker-driven cancer therapy: existing tools and remaining gaps.Discov Med. 2014 Feb;17(92):101-14. Discov Med. 2014. PMID: 24534473 Free PMC article. Review.
-
Moving pan-cancer studies from basic research toward the clinic.Nat Cancer. 2021 Sep;2(9):879-890. doi: 10.1038/s43018-021-00250-4. Epub 2021 Sep 16. Nat Cancer. 2021. PMID: 35121865 Review.
-
Computational Approaches to Prioritize Cancer Driver Missense Mutations.Int J Mol Sci. 2018 Jul 20;19(7):2113. doi: 10.3390/ijms19072113. Int J Mol Sci. 2018. PMID: 30037003 Free PMC article. Review.
References
Publication types
MeSH terms
Grants and funding
- P30 CA016672/CA/NCI NIH HHS/United States
- R01 CA172652/CA/NCI NIH HHS/United States
- CA143883/CA/NCI NIH HHS/United States
- 1U01CA180964/CA/NCI NIH HHS/United States
- P50 CA083639/CA/NCI NIH HHS/United States
- 1R01CA172652/CA/NCI NIH HHS/United States
- CA168394/CA/NCI NIH HHS/United States
- CA083639/CA/NCI NIH HHS/United States
- UL1 TR000371/TR/NCATS NIH HHS/United States
- P50 CA098258/CA/NCI NIH HHS/United States
- U01 CA180964/CA/NCI NIH HHS/United States
- U01 CA168394/CA/NCI NIH HHS/United States
- UL1TR000371/TR/NCATS NIH HHS/United States
- U24 CA143883/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials

