Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice
- PMID: 24204315
- PMCID: PMC3812087
- DOI: 10.1371/journal.pgen.1003913
Loss of miR-10a activates lpo and collaborates with activated Wnt signaling in inducing intestinal neoplasia in female mice
Abstract
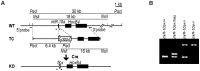
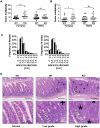
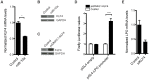
miRNAs are small regulatory RNAs that, due to their considerable potential to target a wide range of mRNAs, are implicated in essentially all biological process, including cancer. miR-10a is particularly interesting considering its conserved location in the Hox cluster of developmental regulators. A role for this microRNA has been described in developmental regulation as well as for various cancers. However, previous miR-10a studies are exclusively based on transient knockdowns of this miRNA and to extensively study miR-10a loss we have generated a miR-10a knock out mouse. Here we show that, in the Apc(min) mouse model of intestinal neoplasia, female miR-10a deficient mice develop significantly more adenomas than miR-10(+/+) and male controls. We further found that Lpo is extensively upregulated in the intestinal epithelium of mice deprived of miR-10a. Using in vitro assays, we demonstrate that the primary miR-10a target KLF4 can upregulate transcription of Lpo, whereas siRNA knockdown of KLF4 reduces LPO levels in HCT-116 cells. Furthermore, Klf4 is upregulated in the intestines of miR-10a knockout mice. Lpo has previously been shown to have the capacity to oxidize estrogens into potent depurinating mutagens, creating an instable genomic environment that can cause initiation of cancer. Therefore, we postulate that Lpo upregulation in the intestinal epithelium of miR-10a deficient mice together with the predominant abundance of estrogens in female animals mainly accounts for the sex-related cancer phenotype we observed. This suggests that miR-10a could be used as a potent diagnostic marker for discovering groups of women that are at high risk of developing colorectal carcinoma, which today is one of the leading causes of cancer-related deaths.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


Similar articles
-
miR-34a and miR-34b/c Suppress Intestinal Tumorigenesis.Cancer Res. 2017 May 15;77(10):2746-2758. doi: 10.1158/0008-5472.CAN-16-2183. Epub 2017 Mar 31. Cancer Res. 2017. PMID: 28363996
-
Tensin4 (TNS4) is upregulated by Wnt signalling in adenomas in multiple intestinal neoplasia (Min) mice.Int J Exp Pathol. 2020 Jun;101(3-4):80-86. doi: 10.1111/iep.12352. Epub 2020 Jun 22. Int J Exp Pathol. 2020. PMID: 32567731 Free PMC article.
-
miR-10a rejuvenates aged human mesenchymal stem cells and improves heart function after myocardial infarction through KLF4.Stem Cell Res Ther. 2018 May 30;9(1):151. doi: 10.1186/s13287-018-0895-0. Stem Cell Res Ther. 2018. PMID: 29848383 Free PMC article.
-
miR-10a restores human mesenchymal stem cell differentiation by repressing KLF4.J Cell Physiol. 2013 Dec;228(12):2324-36. doi: 10.1002/jcp.24402. J Cell Physiol. 2013. PMID: 23696417 Free PMC article.
-
Haploinsufficiency of Krüppel-like factor 4 promotes adenomatous polyposis coli dependent intestinal tumorigenesis.Cancer Res. 2007 Aug 1;67(15):7147-54. doi: 10.1158/0008-5472.CAN-07-1302. Cancer Res. 2007. PMID: 17671182 Free PMC article.
Cited by
-
G9a-dependent histone methylation can be induced in G1 phase of cell cycle.Sci Rep. 2019 Jan 30;9(1):956. doi: 10.1038/s41598-018-37507-5. Sci Rep. 2019. PMID: 30700744 Free PMC article.
-
Transcriptomic profiling in human mesangial cells using patient-derived lupus autoantibodies identified miR-10a as a potential regulator of IL8.Sci Rep. 2017 Nov 6;7(1):14517. doi: 10.1038/s41598-017-15160-8. Sci Rep. 2017. PMID: 29109423 Free PMC article.
-
Ablation of miR-10b Suppresses Oncogene-Induced Mammary Tumorigenesis and Metastasis and Reactivates Tumor-Suppressive Pathways.Cancer Res. 2016 Nov 1;76(21):6424-6435. doi: 10.1158/0008-5472.CAN-16-1571. Epub 2016 Aug 28. Cancer Res. 2016. PMID: 27569213 Free PMC article.
-
HOXs and lincRNAs: Two sides of the same coin.Sci Adv. 2016 Jan 29;2(1):e1501402. doi: 10.1126/sciadv.1501402. eCollection 2016 Jan. Sci Adv. 2016. PMID: 27034976 Free PMC article. Review.
-
Outside the coding genome, mammalian microRNAs confer structural and functional complexity.Sci Signal. 2015 Mar 17;8(368):re2. doi: 10.1126/scisignal.2005813. Sci Signal. 2015. PMID: 25783159 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

