Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy
- PMID: 24191055
- PMCID: PMC3845193
- DOI: 10.1073/pnas.1319280110
Dysregulation of synaptogenesis genes antecedes motor neuron pathology in spinal muscular atrophy
Abstract
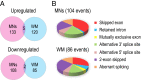
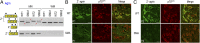
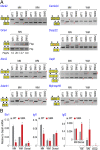
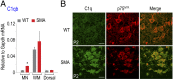
The motor neuron (MN) degenerative disease, spinal muscular atrophy (SMA) is caused by deficiency of SMN (survival motor neuron), a ubiquitous and indispensable protein essential for biogenesis of snRNPs, key components of pre-mRNA processing. However, SMA's hallmark MN pathology, including neuromuscular junction (NMJ) disruption and sensory-motor circuitry impairment, remains unexplained. Toward this end, we used deep RNA sequencing (RNA-seq) to determine if there are any transcriptome changes in MNs and surrounding spinal cord glial cells (white matter, WM) microdissected from SMN-deficient SMA mouse model at presymptomatic postnatal day 1 (P1), before detectable MN pathology (P4-P5). The RNA-seq results, previously unavailable for SMA at any stage, revealed cell-specific selective mRNA dysregulations (~300 of 11,000 expressed genes in each, MN and WM), many of which are known to impair neurons. Remarkably, these dysregulations include complete skipping of agrin's Z exons, critical for NMJ maintenance, strong up-regulation of synapse pruning-promoting complement factor C1q, and down-regulation of Etv1/ER81, a transcription factor required for establishing sensory-motor circuitry. We propose that dysregulation of such specific MN synaptogenesis genes, compounded by many additional transcriptome abnormalities in MNs and WM, link SMN deficiency to SMA's signature pathology.
Keywords: C1q complex; Z+ (neuronal) agrin; transcriptome perturbations.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Survival motor neuron protein in motor neurons determines synaptic integrity in spinal muscular atrophy.J Neurosci. 2012 Jun 20;32(25):8703-15. doi: 10.1523/JNEUROSCI.0204-12.2012. J Neurosci. 2012. PMID: 22723710 Free PMC article.
-
Motor neuronal repletion of the NMJ organizer, Agrin, modulates the severity of the spinal muscular atrophy disease phenotype in model mice.Hum Mol Genet. 2017 Jul 1;26(13):2377-2385. doi: 10.1093/hmg/ddx124. Hum Mol Genet. 2017. PMID: 28379354 Free PMC article.
-
Synaptic defects in spinal muscular atrophy animal models.Dev Neurobiol. 2012 Jan;72(1):126-33. doi: 10.1002/dneu.20912. Dev Neurobiol. 2012. PMID: 21567981 Review.
-
Transcriptome profiling of spinal muscular atrophy motor neurons derived from mouse embryonic stem cells.PLoS One. 2014 Sep 5;9(9):e106818. doi: 10.1371/journal.pone.0106818. eCollection 2014. PLoS One. 2014. PMID: 25191843 Free PMC article.
-
The contribution of mouse models to understanding the pathogenesis of spinal muscular atrophy.Dis Model Mech. 2011 Jul;4(4):457-67. doi: 10.1242/dmm.007245. Dis Model Mech. 2011. PMID: 21708901 Free PMC article. Review.
Cited by
-
Spinal muscular atrophy: journeying from bench to bedside.Neurotherapeutics. 2014 Oct;11(4):786-95. doi: 10.1007/s13311-014-0293-y. Neurotherapeutics. 2014. PMID: 24990202 Free PMC article. Review.
-
Critical Roles of Dual-Specificity Phosphatases in Neuronal Proteostasis and Neurological Diseases.Int J Mol Sci. 2017 Sep 13;18(9):1963. doi: 10.3390/ijms18091963. Int J Mol Sci. 2017. PMID: 28902166 Free PMC article. Review.
-
The SMN Complex at the Crossroad between RNA Metabolism and Neurodegeneration.Int J Mol Sci. 2023 Jan 23;24(3):2247. doi: 10.3390/ijms24032247. Int J Mol Sci. 2023. PMID: 36768569 Free PMC article. Review.
-
Hyperexcitability precedes motoneuron loss in the Smn2B/- mouse model of spinal muscular atrophy.J Neurophysiol. 2019 Oct 1;122(4):1297-1311. doi: 10.1152/jn.00652.2018. Epub 2019 Jul 31. J Neurophysiol. 2019. PMID: 31365319 Free PMC article.
-
Altered mitochondrial bioenergetics are responsible for the delay in Wallerian degeneration observed in neonatal mice.Neurobiol Dis. 2019 Oct;130:104496. doi: 10.1016/j.nbd.2019.104496. Epub 2019 Jun 6. Neurobiol Dis. 2019. PMID: 31176719 Free PMC article.
References
-
- Cifuentes-Diaz C, Frugier T, Melki J. Spinal muscular atrophy. Semin Pediatr Neurol. 2002;9(2):145–150. - PubMed
-
- Wee CD, Kong L, Sumner CJ. The genetics of spinal muscular atrophies. Curr Opin Neurol. 2010;23(5):450–458. - PubMed
-
- Lefebvre S, et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80(1):155–165. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

