Molecular determinants of PI3Kγ-mediated activation downstream of G-protein-coupled receptors (GPCRs)
- PMID: 24190998
- PMCID: PMC3839701
- DOI: 10.1073/pnas.1304801110
Molecular determinants of PI3Kγ-mediated activation downstream of G-protein-coupled receptors (GPCRs)
Abstract
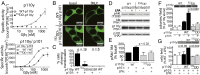
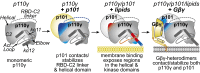
Phosphoinositide 3-kinase gamma (PI3Kγ) has profound roles downstream of G-protein-coupled receptors in inflammation, cardiac function, and tumor progression. To gain insight into how the enzyme's activity is shaped by association with its p101 adaptor subunit, lipid membranes, and Gβγ heterodimers, we mapped these regulatory interactions using hydrogen-deuterium exchange mass spectrometry. We identify residues in both the p110γ and p101 subunits that contribute critical interactions with Gβγ heterodimers, leading to PI3Kγ activation. Mutating Gβγ-interaction sites of either p110γ or p101 ablates G-protein-coupled receptor-mediated signaling to p110γ/p101 in cells and severely affects chemotaxis and cell transformation induced by PI3Kγ overexpression. Hydrogen-deuterium exchange mass spectrometry shows that association with the p101 regulatory subunit causes substantial protection of the RBD-C2 linker as well as the helical domain of p110γ. Lipid interaction massively exposes that same helical site, which is then stabilized by Gβγ. Membrane-elicited conformational change of the helical domain could help prepare the enzyme for Gβγ binding. Our studies and others identify the helical domain of the class I PI3Ks as a hub for diverse regulatory interactions that include the p101, p87 (also known as p84), and p85 adaptor subunits; Rab5 and Gβγ heterodimers; and the β-adrenergic receptor kinase.
Keywords: HDX-MS; PIK3CG; PIK3R5; PIP3; oncogene.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Different inhibition of Gβγ-stimulated class IB phosphoinositide 3-kinase (PI3K) variants by a monoclonal antibody. Specific function of p101 as a Gβγ-dependent regulator of PI3Kγ enzymatic activity.Biochem J. 2015 Jul 1;469(1):59-69. doi: 10.1042/BJ20150099. Biochem J. 2015. PMID: 26173259 Free PMC article.
-
Molecular basis for differential activation of p101 and p84 complexes of PI3Kγ by Ras and GPCRs.Cell Rep. 2023 Mar 28;42(3):112172. doi: 10.1016/j.celrep.2023.112172. Epub 2023 Feb 26. Cell Rep. 2023. PMID: 36842083 Free PMC article.
-
Ras is an indispensable coregulator of the class IB phosphoinositide 3-kinase p87/p110gamma.Proc Natl Acad Sci U S A. 2009 Dec 1;106(48):20312-7. doi: 10.1073/pnas.0905506106. Epub 2009 Nov 11. Proc Natl Acad Sci U S A. 2009. PMID: 19906996 Free PMC article.
-
Function, Regulation and Biological Roles of PI3Kγ Variants.Biomolecules. 2019 Aug 30;9(9):427. doi: 10.3390/biom9090427. Biomolecules. 2019. PMID: 31480354 Free PMC article. Review.
-
PI3K class IB pathway in neutrophils.Sci STKE. 2007 Oct 9;2007(407):cm3. doi: 10.1126/stke.4072007cm3. Sci STKE. 2007. PMID: 17925574 Review.
Cited by
-
Beyond PI3Ks: targeting phosphoinositide kinases in disease.Nat Rev Drug Discov. 2023 May;22(5):357-386. doi: 10.1038/s41573-022-00582-5. Epub 2022 Nov 14. Nat Rev Drug Discov. 2023. PMID: 36376561 Free PMC article. Review.
-
Akt-ing Up Just About Everywhere: Compartment-Specific Akt Activation and Function in Receptor Tyrosine Kinase Signaling.Front Cell Dev Biol. 2019 May 3;7:70. doi: 10.3389/fcell.2019.00070. eCollection 2019. Front Cell Dev Biol. 2019. PMID: 31131274 Free PMC article. Review.
-
Activation of Phospholipase C β by Gβγ and Gαq Involves C-Terminal Rearrangement to Release Autoinhibition.Structure. 2020 Jul 7;28(7):810-819.e5. doi: 10.1016/j.str.2020.04.012. Epub 2020 May 12. Structure. 2020. PMID: 32402248 Free PMC article.
-
Distinct roles for phosphoinositide 3-kinases γ and δ in malignant B cell migration.Leukemia. 2018 Sep;32(9):1958-1969. doi: 10.1038/s41375-018-0012-5. Epub 2018 Jan 31. Leukemia. 2018. PMID: 29479062 Free PMC article.
-
PI3K inhibitors in thrombosis and cardiovascular disease.Clin Transl Med. 2020 Jan 31;9(1):8. doi: 10.1186/s40169-020-0261-6. Clin Transl Med. 2020. PMID: 32002690 Free PMC article. Review.
References
-
- Sasaki T, et al. Function of PI3Kgamma in thymocyte development, T cell activation, and neutrophil migration. Science. 2000;287(5455):1040–1046. - PubMed
-
- Hirsch E, et al. Central role for G protein-coupled phosphoinositide 3-kinase gamma in inflammation. Science. 2000;287(5455):1049–1053. - PubMed
-
- Li Z, et al. Roles of PLC-beta2 and -beta3 and PI3Kgamma in chemoattractant-mediated signal transduction. Science. 2000;287(5455):1046–1049. - PubMed
-
- Laffargue M, et al. Phosphoinositide 3-kinase gamma is an essential amplifier of mast cell function. Immunity. 2002;16(3):441–451. - PubMed
-
- Martin EL, et al. Phosphoinositide-3 kinase gamma activity contributes to sepsis and organ damage by altering neutrophil recruitment. Am J Respir Crit Care Med. 2010;182(6):762–773. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

