Enhanced autophagy ameliorates cardiac proteinopathy
- PMID: 24177425
- PMCID: PMC3859422
- DOI: 10.1172/JCI70877
Enhanced autophagy ameliorates cardiac proteinopathy
Abstract
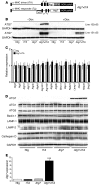
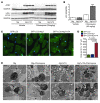
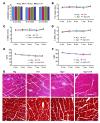
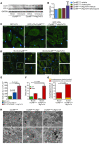
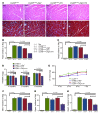
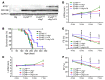
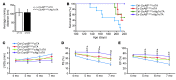
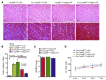
Basal autophagy is a crucial mechanism in cellular homeostasis, underlying both normal cellular recycling and the clearance of damaged or misfolded proteins, organelles and aggregates. We showed here that enhanced levels of autophagy induced by either autophagic gene overexpression or voluntary exercise ameliorated desmin-related cardiomyopathy (DRC). To increase levels of basal autophagy, we generated an inducible Tg mouse expressing autophagy-related 7 (Atg7), a critical and rate-limiting autophagy protein. Hearts from these mice had enhanced autophagy, but normal morphology and function. We crossed these mice with CryABR120G mice, a model of DRC in which autophagy is significantly attenuated in the heart, to test the functional significance of autophagy activation in a proteotoxic model of heart failure. Sustained Atg7-induced autophagy in the CryABR120G hearts decreased interstitial fibrosis, ameliorated ventricular dysfunction, decreased cardiac hypertrophy, reduced intracellular aggregates and prolonged survival. To determine whether different methods of autophagy upregulation have additive or even synergistic benefits, we subjected the autophagy-deficient CryABR120G mice and the Atg7-crossed CryABR120G mice to voluntary exercise, which also upregulates autophagy. The entire exercised Atg7-crossed CryABR120G cohort survived to 7 months. These findings suggest that activating autophagy may be a viable therapeutic strategy for improving cardiac performance under proteotoxic conditions.
Figures

Similar articles
-
Autophagy and p62 in cardiac proteinopathy.Circ Res. 2011 Jul 22;109(3):296-308. doi: 10.1161/CIRCRESAHA.111.244707. Epub 2011 Jun 9. Circ Res. 2011. PMID: 21659648 Free PMC article.
-
Atg7 induces basal autophagy and rescues autophagic deficiency in CryABR120G cardiomyocytes.Circ Res. 2011 Jul 8;109(2):151-60. doi: 10.1161/CIRCRESAHA.110.237339. Epub 2011 May 26. Circ Res. 2011. PMID: 21617129 Free PMC article.
-
Autophagy in desmin-related cardiomyopathy: thoughts at the halfway point.Autophagy. 2010 Jul;6(5):665-6. doi: 10.1161/CIRCRESAHA.109.212639. Autophagy. 2010. PMID: 20523125
-
Morphological characteristics in diabetic cardiomyopathy associated with autophagy.J Cardiol. 2021 Jan;77(1):30-40. doi: 10.1016/j.jjcc.2020.05.009. Epub 2020 Sep 6. J Cardiol. 2021. PMID: 32907780 Review.
-
Amplification of lipotoxic cardiomyopathy in the VDR gene knockout mouse.J Steroid Biochem Mol Biol. 2016 Nov;164:292-298. doi: 10.1016/j.jsbmb.2015.09.034. Epub 2015 Sep 30. J Steroid Biochem Mol Biol. 2016. PMID: 26429397 Review.
Cited by
-
Targeting Protein Kinase G to Treat Cardiac Proteotoxicity.Front Physiol. 2020 Jul 28;11:858. doi: 10.3389/fphys.2020.00858. eCollection 2020. Front Physiol. 2020. PMID: 32848832 Free PMC article. Review.
-
Hyperactive mTORC1/4EBP1 signaling dysregulates proteostasis and accelerates cardiac aging.Geroscience. 2024 Oct 9. doi: 10.1007/s11357-024-01368-w. Online ahead of print. Geroscience. 2024. PMID: 39379739
-
The Role of Autophagy in Osteoarthritic Cartilage.Biomolecules. 2022 Sep 23;12(10):1357. doi: 10.3390/biom12101357. Biomolecules. 2022. PMID: 36291565 Free PMC article. Review.
-
Self-eating: friend or foe? The emerging role of autophagy in fibrotic diseases.Theranostics. 2020 Jun 29;10(18):7993-8017. doi: 10.7150/thno.47826. eCollection 2020. Theranostics. 2020. PMID: 32724454 Free PMC article. Review.
-
FYCO1 Regulates Cardiomyocyte Autophagy and Prevents Heart Failure Due to Pressure Overload In Vivo.JACC Basic Transl Sci. 2021 Mar 17;6(4):365-380. doi: 10.1016/j.jacbts.2021.01.001. eCollection 2021 Apr. JACC Basic Transl Sci. 2021. PMID: 33997522 Free PMC article.
References
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

