Evolutionary and ontogenetic changes in RNA editing in human, chimpanzee, and macaque brains
- PMID: 24152549
- PMCID: PMC3884655
- DOI: 10.1261/rna.039206.113
Evolutionary and ontogenetic changes in RNA editing in human, chimpanzee, and macaque brains
Abstract
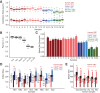
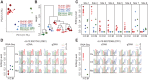
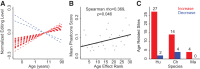
Adenosine-to-inosine (A-to-I) substitutions are the most common type of RNA editing in mammals. A-to-I RNA editing is particularly widespread in the brain and is known to play important roles in neuronal functions. In this study we investigated RNA-editing changes during human brain development and maturation, as well as evolutionary conservation of RNA-editing patterns across primates. We used high-throughput transcriptome sequencing (RNA-seq) to quantify the RNA-editing levels and assess ontogenetic dynamics of RNA editing at more than 8000 previously annotated exonic A-to-I RNA-editing sites in two brain regions--prefrontal cortex and cerebellum--of humans, chimpanzees, and rhesus macaques. We observed substantial conservation of RNA-editing levels between the brain regions, as well as among the three primate species. Evolutionary changes in RNA editing were nonetheless evident, with 40% of the annotated editing sites studied showing divergent editing levels among the three species and 16.5% of sites displaying statistically significant human-specific editing patterns. Across lifespan, we observed an increase of the RNA-editing level with advanced age in both brain regions of all three primate species.
Keywords: RNA editing; aging; brain; chimpanzee; evolution; human.
Figures
Similar articles
-
Predominant patterns of splicing evolution on human, chimpanzee and macaque evolutionary lineages.Hum Mol Genet. 2018 Apr 15;27(8):1474-1485. doi: 10.1093/hmg/ddy058. Hum Mol Genet. 2018. PMID: 29452398
-
Adenosine-to-inosine RNA editing shapes transcriptome diversity in primates.Proc Natl Acad Sci U S A. 2010 Jul 6;107(27):12174-9. doi: 10.1073/pnas.1006183107. Epub 2010 Jun 21. Proc Natl Acad Sci U S A. 2010. PMID: 20566853 Free PMC article.
-
Species-Specific Changes in a Primate Transcription Factor Network Provide Insights into the Molecular Evolution of the Primate Prefrontal Cortex.Genome Biol Evol. 2018 Aug 1;10(8):2023-2036. doi: 10.1093/gbe/evy149. Genome Biol Evol. 2018. PMID: 30059966 Free PMC article.
-
Letter from the editor: Adenosine-to-inosine RNA editing in Alu repeats in the human genome.EMBO Rep. 2005 Sep;6(9):831-5. doi: 10.1038/sj.embor.7400507. EMBO Rep. 2005. PMID: 16138094 Free PMC article. Review.
-
ALU A-to-I RNA Editing: Millions of Sites and Many Open Questions.Methods Mol Biol. 2021;2181:149-162. doi: 10.1007/978-1-0716-0787-9_9. Methods Mol Biol. 2021. PMID: 32729079 Review.
Cited by
-
Regulatory factors governing adenosine-to-inosine (A-to-I) RNA editing.Biosci Rep. 2015 Mar 31;35(2):e00182. doi: 10.1042/BSR20140190. Biosci Rep. 2015. PMID: 25662729 Free PMC article. Review.
-
Comparative Hippocampal Synaptic Proteomes of Rodents and Primates: Differences in Neuroplasticity-Related Proteins.Front Mol Neurosci. 2018 Oct 2;11:364. doi: 10.3389/fnmol.2018.00364. eCollection 2018. Front Mol Neurosci. 2018. PMID: 30333727 Free PMC article.
-
A-to-I RNA editing - immune protector and transcriptome diversifier.Nat Rev Genet. 2018 Aug;19(8):473-490. doi: 10.1038/s41576-018-0006-1. Nat Rev Genet. 2018. PMID: 29692414 Review.
-
Evolutionary dynamics of coding and non-coding transcriptomes.Nat Rev Genet. 2014 Nov;15(11):734-48. doi: 10.1038/nrg3802. Epub 2014 Oct 9. Nat Rev Genet. 2014. PMID: 25297727 Review.
-
Analytical tools and current challenges in the modern era of neuroepigenomics.Nat Neurosci. 2014 Nov;17(11):1476-90. doi: 10.1038/nn.3816. Epub 2014 Oct 28. Nat Neurosci. 2014. PMID: 25349914 Free PMC article. Review.
References
-
- Baggerly KA, Deng L, Morris JS, Aldaz CM. 2003. Differential expression in SAGE: Accounting for normal between-library variation. Bioinformatics 19: 1477–1483. - PubMed
-
- Bass BL, Weintraub H. 1988. An unwinding activity that covalently modifies its double-stranded RNA substrate. Cell 55: 1089–1098. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
