A potential diagnostic marker for ovarian cancer: Involvement of the histone acetyltransferase, human males absent on the first
- PMID: 24137335
- PMCID: PMC3789056
- DOI: 10.3892/ol.2013.1380
A potential diagnostic marker for ovarian cancer: Involvement of the histone acetyltransferase, human males absent on the first
Abstract
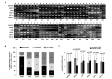
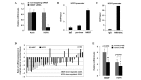
Human males absent on the first (hMOF), a human ortholog of the Drosophila MOF protein, is responsible for histone H4 lysine 16 (H4K16) acetylation in human cells. The depletion of hMOF leads to a global reduction in histone H4K16 acetylation in human cells, genomic instability, cell cycle defects, reduced transcription of certain genes, defective DNA damage repair and early embryonic lethality. Studies have shown that abnormal hMOF gene expression is involved in a number of primary cancers. The present study examined the involvement of hMOF expression and histone H4K16 acetylation in clinically diagnosed primary ovarian cancer tissues. Clinically diagnosed frozen primary ovarian cancer tissues were used for polymerase chain reaction (PCR), quantitative PCR (qPCR), western blotting and immunohistochemical staining approaches. A PCR analysis of mRNA expression in 47 samples revealed a downregulation of hMOF mRNA in 81% of patients, whereas only 13% of patients demonstrated upregulation. qPCR was used to validate the frequent downregulation of hMOF expression in the primary ovarian cancer tissues. As expected, the analysis of hMOF expression in 57 samples revealed that hMOF mRNA expression was significantly downregulated (>2-fold decrease) in 65% of patients, while a <2-fold reduction of hMOF was observed in 10.5% of patients. Furthermore, the expression of hMOF-regulated human leukocyte antigen (HLA) complex 5, (HCP5), was also found to be downregulated in >87% of patients with a decrease in hMOF. hMOF and its regulated gene, HCP5, are frequently downregulated in human ovarian cancer, suggesting that hMOF may be involved in the pathogenesis of the disease.
Keywords: histone acetyltransferase; human males absent on the first; ovarian cancer.
Figures


Similar articles
-
Epigenetic change in kidney tumor: downregulation of histone acetyltransferase MYST1 in human renal cell carcinoma.J Exp Clin Cancer Res. 2013 Feb 9;32(1):8. doi: 10.1186/1756-9966-32-8. J Exp Clin Cancer Res. 2013. PMID: 23394073 Free PMC article.
-
The histone acetyltransferase hMOF is frequently downregulated in primary breast carcinoma and medulloblastoma and constitutes a biomarker for clinical outcome in medulloblastoma.Int J Cancer. 2008 Mar 15;122(6):1207-13. doi: 10.1002/ijc.23283. Int J Cancer. 2008. PMID: 18058815
-
Correlation of low expression of hMOF with clinicopathological features of colorectal carcinoma, gastric cancer and renal cell carcinoma.Int J Oncol. 2014 Apr;44(4):1207-14. doi: 10.3892/ijo.2014.2266. Epub 2014 Jan 21. Int J Oncol. 2014. PMID: 24452485
-
Males absent on the first (MOF): from flies to humans.Oncogene. 2007 Aug 13;26(37):5385-94. doi: 10.1038/sj.onc.1210607. Oncogene. 2007. PMID: 17694080 Review.
-
The Functional Analysis of Histone Acetyltransferase MOF in Tumorigenesis.Int J Mol Sci. 2016 Jan 14;17(1):99. doi: 10.3390/ijms17010099. Int J Mol Sci. 2016. PMID: 26784169 Free PMC article. Review.
Cited by
-
LncRNA HCP5-Encoded Protein Regulates Ferroptosis to Promote the Progression of Triple-Negative Breast Cancer.Cancers (Basel). 2023 Mar 21;15(6):1880. doi: 10.3390/cancers15061880. Cancers (Basel). 2023. PMID: 36980766 Free PMC article.
-
The multifaceted role of lysine acetylation in cancer: prognostic biomarker and therapeutic target.Oncotarget. 2016 Aug 23;7(34):55789-55810. doi: 10.18632/oncotarget.10048. Oncotarget. 2016. PMID: 27322556 Free PMC article. Review.
-
Epigenetics of Most Aggressive Solid Tumors: Pathways, Targets and Treatments.Cancers (Basel). 2021 Jun 27;13(13):3209. doi: 10.3390/cancers13133209. Cancers (Basel). 2021. PMID: 34198989 Free PMC article. Review.
-
Knockdown of Long Non-Coding RNA HCP5 Increases Radiosensitivity Through Cellular Senescence by Regulating microRNA-128 in Gliomas.Cancer Manag Res. 2021 May 7;13:3723-3737. doi: 10.2147/CMAR.S301333. eCollection 2021. Cancer Manag Res. 2021. PMID: 33994812 Free PMC article.
-
Construction for Long Non-Coding RNA (lncRNA)-Associated Competing Endogenous RNA (ceRNA) Network in Human Retinal Detachment (RD) with Proliferative Vitreoretinopathy (PVR).Med Sci Monit. 2020 Feb 27;26:e919871. doi: 10.12659/MSM.919871. Med Sci Monit. 2020. PMID: 32103829 Free PMC article.
References
-
- Mendjan S, Taipale M, Kind J, Holz H, Gebhardt P, Schelder M, Vermeulen M, Buscaino A, Duncan K, Mueller J, Wilm M, Stunnenberg HG, Saumweber H, Akhtar A. Nuclear pore components are involved in the transcriptional regulation of dosage compensation in Drosophila. Mol Cell. 2006;21:811–823. - PubMed
-
- Carrozza MJ, Utley RT, Workman JL, Côté J. The diverse functions of histone acetyltransferase complexes. Trends Genet. 2003;19:321–329. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
