De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts
- PMID: 24106303
- PMCID: PMC3808619
- DOI: 10.1073/pnas.1306262110
De novo identification of VRC01 class HIV-1-neutralizing antibodies by next-generation sequencing of B-cell transcripts
Abstract
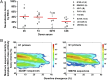
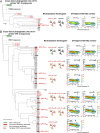
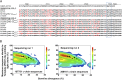
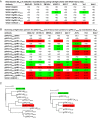
Next-generation sequencing of antibody transcripts provides a wealth of data, but the ability to identify function-specific antibodies solely on the basis of sequence has remained elusive. We previously characterized the VRC01 class of antibodies, which target the CD4-binding site on gp120, appear in multiple donors, and broadly neutralize HIV-1. Antibodies of this class have developmental commonalities, but typically share only ∼50% amino acid sequence identity among different donors. Here we apply next-generation sequencing to identify VRC01 class antibodies in a new donor, C38, directly from B cell transcript sequences. We first tested a lineage rank approach, but this was unsuccessful, likely because VRC01 class antibody sequences were not highly prevalent in this donor. We next identified VRC01 class heavy chains through a phylogenetic analysis that included thousands of sequences from C38 and a few known VRC01 class sequences from other donors. This "cross-donor analysis" yielded heavy chains with little sequence homology to previously identified VRC01 class heavy chains. Nonetheless, when reconstituted with the light chain from VRC01, half of the heavy chain chimeric antibodies showed substantial neutralization potency and breadth. We then identified VRC01 class light chains through a five-amino-acid sequence motif necessary for VRC01 light chain recognition. From over a million light chain sequences, we identified 13 candidate VRC01 class members. Pairing of these light chains with the phylogenetically identified C38 heavy chains yielded functional antibodies that effectively neutralized HIV-1. Bioinformatics analysis can thus directly identify functional HIV-1-neutralizing antibodies of the VRC01 class from a sequenced antibody repertoire.
Keywords: DNA sequencing; antibodyomics; cross-donor phylogenetic analysis; humoral immune response; sequence signature.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Focused evolution of HIV-1 neutralizing antibodies revealed by structures and deep sequencing.Science. 2011 Sep 16;333(6049):1593-602. doi: 10.1126/science.1207532. Epub 2011 Aug 11. Science. 2011. PMID: 21835983 Free PMC article.
-
Mining the antibodyome for HIV-1-neutralizing antibodies with next-generation sequencing and phylogenetic pairing of heavy/light chains.Proc Natl Acad Sci U S A. 2013 Apr 16;110(16):6470-5. doi: 10.1073/pnas.1219320110. Epub 2013 Mar 27. Proc Natl Acad Sci U S A. 2013. PMID: 23536288 Free PMC article.
-
Multidonor analysis reveals structural elements, genetic determinants, and maturation pathway for HIV-1 neutralization by VRC01-class antibodies.Immunity. 2013 Aug 22;39(2):245-58. doi: 10.1016/j.immuni.2013.04.012. Epub 2013 Aug 1. Immunity. 2013. PMID: 23911655 Free PMC article.
-
Elicitation of HIV-1-neutralizing antibodies against the CD4-binding site.Curr Opin HIV AIDS. 2013 Sep;8(5):382-92. doi: 10.1097/COH.0b013e328363a90e. Curr Opin HIV AIDS. 2013. PMID: 23924998 Free PMC article. Review.
-
A matrix of structure-based designs yields improved VRC01-class antibodies for HIV-1 therapy and prevention.MAbs. 2021 Jan-Dec;13(1):1946918. doi: 10.1080/19420862.2021.1946918. MAbs. 2021. PMID: 34328065 Free PMC article. Review.
Cited by
-
Genetic and structural insights into broad neutralization of hepatitis C virus by human VH1-69 antibodies.Sci Adv. 2019 Jan 2;5(1):eaav1882. doi: 10.1126/sciadv.aav1882. eCollection 2019 Jan. Sci Adv. 2019. PMID: 30613781 Free PMC article.
-
Practical guidelines for B-cell receptor repertoire sequencing analysis.Genome Med. 2015 Nov 20;7:121. doi: 10.1186/s13073-015-0243-2. Genome Med. 2015. PMID: 26589402 Free PMC article. Review.
-
The evolution within us.Philos Trans R Soc Lond B Biol Sci. 2015 Sep 5;370(1676):20140235. doi: 10.1098/rstb.2014.0235. Philos Trans R Soc Lond B Biol Sci. 2015. PMID: 26194749 Free PMC article. Review.
-
HIV broadly neutralizing antibody targets.Curr Opin HIV AIDS. 2015 May;10(3):135-43. doi: 10.1097/COH.0000000000000153. Curr Opin HIV AIDS. 2015. PMID: 25760932 Free PMC article. Review.
-
Differences in Allelic Frequency and CDRH3 Region Limit the Engagement of HIV Env Immunogens by Putative VRC01 Neutralizing Antibody Precursors.Cell Rep. 2016 Nov 1;17(6):1560-1570. doi: 10.1016/j.celrep.2016.10.017. Cell Rep. 2016. PMID: 27806295 Free PMC article.
References
-
- Colman PM. Structure of antibody-antigen complexes: Implications for immune recognition. Adv Immunol. 1988;43:99–132. - PubMed
-
- Padlan EA. Structural basis for the specificity of antibody-antigen reactions and structural mechanisms for the diversification of antigen-binding specificities. Q Rev Biophys. 1977;10(1):35–65. - PubMed
-
- Wilson IA, Rini JM, Fremont DH, Fieser GG, Stura EA. X-ray crystallographic analysis of free and antigen-complexed Fab fragments to investigate structural basis of immune recognition. Methods Enzymol. 1991;203:153–176. - PubMed
-
- Schueler-Furman O, Wang C, Bradley P, Misura K, Baker D. Progress in modeling of protein structures and interactions. Science. 2005;310(5748):638–642. - PubMed
-
- Kolodny R, Petrey D, Honig B. Protein structure comparison: Implications for the nature of ‘fold space’, and structure and function prediction. Curr Opin Struct Biol. 2006;16(3):393–398. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

