NaviCell: a web-based environment for navigation, curation and maintenance of large molecular interaction maps
- PMID: 24099179
- PMCID: PMC3851986
- DOI: 10.1186/1752-0509-7-100
NaviCell: a web-based environment for navigation, curation and maintenance of large molecular interaction maps
Abstract
Background: Molecular biology knowledge can be formalized and systematically represented in a computer-readable form as a comprehensive map of molecular interactions. There exist an increasing number of maps of molecular interactions containing detailed and step-wise description of various cell mechanisms. It is difficult to explore these large maps, to organize discussion of their content and to maintain them. Several efforts were recently made to combine these capabilities together in one environment, and NaviCell is one of them.
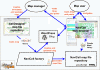
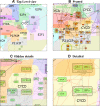
Results: NaviCell is a web-based environment for exploiting large maps of molecular interactions, created in CellDesigner, allowing their easy exploration, curation and maintenance. It is characterized by a combination of three essential features: (1) efficient map browsing based on Google Maps; (2) semantic zooming for viewing different levels of details or of abstraction of the map and (3) integrated web-based blog for collecting community feedback. NaviCell can be easily used by experts in the field of molecular biology for studying molecular entities of interest in the context of signaling pathways and crosstalk between pathways within a global signaling network. NaviCell allows both exploration of detailed molecular mechanisms represented on the map and a more abstract view of the map up to a top-level modular representation. NaviCell greatly facilitates curation, maintenance and updating the comprehensive maps of molecular interactions in an interactive and user-friendly fashion due to an imbedded blogging system.
Conclusions: NaviCell provides user-friendly exploration of large-scale maps of molecular interactions, thanks to Google Maps and WordPress interfaces, with which many users are already familiar. Semantic zooming which is used for navigating geographical maps is adopted for molecular maps in NaviCell, making any level of visualization readable. In addition, NaviCell provides a framework for community-based curation of maps.
Figures



Similar articles
-
NaviCell Web Service for network-based data visualization.Nucleic Acids Res. 2015 Jul 1;43(W1):W560-5. doi: 10.1093/nar/gkv450. Epub 2015 May 9. Nucleic Acids Res. 2015. PMID: 25958393 Free PMC article.
-
Signalling maps in cancer research: construction and data analysis.Database (Oxford). 2018 Jan 1;2018:bay036. doi: 10.1093/database/bay036. Database (Oxford). 2018. PMID: 29688383 Free PMC article.
-
Atlas of Cancer Signalling Network: a systems biology resource for integrative analysis of cancer data with Google Maps.Oncogenesis. 2015 Jul 20;4(7):e160. doi: 10.1038/oncsis.2015.19. Oncogenesis. 2015. PMID: 26192618 Free PMC article.
-
Application of Atlas of Cancer Signalling Network in preclinical studies.Brief Bioinform. 2019 Mar 25;20(2):701-716. doi: 10.1093/bib/bby031. Brief Bioinform. 2019. PMID: 29726961 Review.
-
Integrating Text Mining into the Curation of Disease Maps.Biomolecules. 2022 Sep 10;12(9):1278. doi: 10.3390/biom12091278. Biomolecules. 2022. PMID: 36139119 Free PMC article. Review.
Cited by
-
A guide for developing comprehensive systems biology maps of disease mechanisms: planning, construction and maintenance.Front Bioinform. 2023 Jun 22;3:1197310. doi: 10.3389/fbinf.2023.1197310. eCollection 2023. Front Bioinform. 2023. PMID: 37426048 Free PMC article.
-
Closing the gap between formats for storing layout information in systems biology.Brief Bioinform. 2020 Jul 15;21(4):1249-1260. doi: 10.1093/bib/bbz067. Brief Bioinform. 2020. PMID: 31273380 Free PMC article.
-
Boolean modelling as a logic-based dynamic approach in systems medicine.Comput Struct Biotechnol J. 2022 Jun 17;20:3161-3172. doi: 10.1016/j.csbj.2022.06.035. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 35782730 Free PMC article. Review.
-
BioKC: a collaborative platform for curation and annotation of molecular interactions.Database (Oxford). 2024 Mar 27;2024:baae013. doi: 10.1093/database/baae013. Database (Oxford). 2024. PMID: 38537198 Free PMC article.
-
Comprehensive Map of the Regulated Cell Death Signaling Network: A Powerful Analytical Tool for Studying Diseases.Cancers (Basel). 2020 Apr 17;12(4):990. doi: 10.3390/cancers12040990. Cancers (Basel). 2020. PMID: 32316560 Free PMC article.
References
-
- Barillot E, Calzone L, Hupé P, Vert J-P, Zinovyev A. Computational systems biology of cancer. London, UK: CRC Press; 2012.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

