Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals
- PMID: 24092820
- PMCID: PMC3875855
- DOI: 10.1101/gr.155192.113
Characterizing the genetic basis of transcriptome diversity through RNA-sequencing of 922 individuals
Abstract
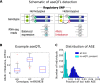
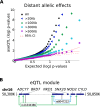
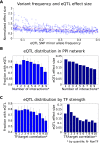
Understanding the consequences of regulatory variation in the human genome remains a major challenge, with important implications for understanding gene regulation and interpreting the many disease-risk variants that fall outside of protein-coding regions. Here, we provide a direct window into the regulatory consequences of genetic variation by sequencing RNA from 922 genotyped individuals. We present a comprehensive description of the distribution of regulatory variation--by the specific expression phenotypes altered, the properties of affected genes, and the genomic characteristics of regulatory variants. We detect variants influencing expression of over ten thousand genes, and through the enhanced resolution offered by RNA-sequencing, for the first time we identify thousands of variants associated with specific phenotypes including splicing and allelic expression. Evaluating the effects of both long-range intra-chromosomal and trans (cross-chromosomal) regulation, we observe modularity in the regulatory network, with three-dimensional chromosomal configuration playing a particular role in regulatory modules within each chromosome. We also observe a significant depletion of regulatory variants affecting central and critical genes, along with a trend of reduced effect sizes as variant frequency increases, providing evidence that purifying selection and buffering have limited the deleterious impact of regulatory variation on the cell. Further, generalizing beyond observed variants, we have analyzed the genomic properties of variants associated with expression and splicing and developed a Bayesian model to predict regulatory consequences of genetic variants, applicable to the interpretation of individual genomes and disease studies. Together, these results represent a critical step toward characterizing the complete landscape of human regulatory variation.
Figures


Similar articles
-
Epistatic selection between coding and regulatory variation in human evolution and disease.Am J Hum Genet. 2011 Sep 9;89(3):459-63. doi: 10.1016/j.ajhg.2011.08.004. Am J Hum Genet. 2011. PMID: 21907014 Free PMC article.
-
Transcriptome and genome sequencing uncovers functional variation in humans.Nature. 2013 Sep 26;501(7468):506-11. doi: 10.1038/nature12531. Epub 2013 Sep 15. Nature. 2013. PMID: 24037378 Free PMC article.
-
Modified penetrance of coding variants by cis-regulatory variation contributes to disease risk.Nat Genet. 2018 Sep;50(9):1327-1334. doi: 10.1038/s41588-018-0192-y. Epub 2018 Aug 20. Nat Genet. 2018. PMID: 30127527 Free PMC article.
-
Impacts of variation in the human genome on gene regulation.J Mol Biol. 2013 Nov 1;425(21):3970-7. doi: 10.1016/j.jmb.2013.07.015. Epub 2013 Jul 16. J Mol Biol. 2013. PMID: 23871684 Review.
-
Interpreting functional effects of coding variants: challenges in proteome-scale prediction, annotation and assessment.Brief Bioinform. 2016 Sep;17(5):841-62. doi: 10.1093/bib/bbv084. Epub 2015 Oct 22. Brief Bioinform. 2016. PMID: 26494363 Free PMC article. Review.
Cited by
-
Schizophrenia interactome with 504 novel protein-protein interactions.NPJ Schizophr. 2016 Apr 27;2:16012. doi: 10.1038/npjschz.2016.12. eCollection 2016. NPJ Schizophr. 2016. PMID: 27336055 Free PMC article.
-
How to Predict Molecular Interactions between Species?Front Microbiol. 2016 Mar 31;7:442. doi: 10.3389/fmicb.2016.00442. eCollection 2016. Front Microbiol. 2016. PMID: 27065992 Free PMC article. Review.
-
Combinatorial and statistical prediction of gene expression from haplotype sequence.Bioinformatics. 2020 Jul 1;36(Suppl_1):i194-i202. doi: 10.1093/bioinformatics/btaa318. Bioinformatics. 2020. PMID: 32657373 Free PMC article.
-
A gene-based association method for mapping traits using reference transcriptome data.Nat Genet. 2015 Sep;47(9):1091-8. doi: 10.1038/ng.3367. Epub 2015 Aug 10. Nat Genet. 2015. PMID: 26258848 Free PMC article.
-
Transcriptomics and chromatin accessibility in multiple African population samples.bioRxiv [Preprint]. 2023 Nov 6:2023.11.04.564839. doi: 10.1101/2023.11.04.564839. bioRxiv. 2023. PMID: 37986808 Free PMC article. Preprint.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
