GRL-04810 and GRL-05010, difluoride-containing nonpeptidic HIV-1 protease inhibitors (PIs) that inhibit the replication of multi-PI-resistant HIV-1 in vitro and possess favorable lipophilicity that may allow blood-brain barrier penetration
- PMID: 24080647
- PMCID: PMC3837900
- DOI: 10.1128/AAC.01420-13
GRL-04810 and GRL-05010, difluoride-containing nonpeptidic HIV-1 protease inhibitors (PIs) that inhibit the replication of multi-PI-resistant HIV-1 in vitro and possess favorable lipophilicity that may allow blood-brain barrier penetration
Abstract
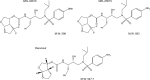
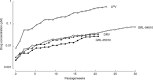
We designed, synthesized, and identified two novel nonpeptidic human immunodeficiency virus type 1 (HIV-1) protease inhibitors (PIs), GRL-04810 and GRL-05010, containing the structure-based designed privileged cyclic ether-derived nonpeptide P2 ligand, bis-tetrahydrofuranylurethane (bis-THF), and a difluoride moiety, both of which are active against the laboratory strain HIV-1LAI (50% effective concentrations [EC50s], 0.0008 and 0.003 μM, respectively) with minimal cytotoxicity (50% cytotoxic concentrations [CC50s], 17.5 and 37.0 μM, respectively, in CD4(+) MT-2 cells). The two compounds were active against multi-PI-resistant clinical HIV-1 variants isolated from patients who had no response to various antiviral regimens. GRL-04810 and GRL-05010 also blocked the infectivity and replication of each of the HIV-1NL4-3 variants selected by up to 5 μM lopinavir (EC50s, 0.03 and 0.03 μM, respectively) and atazanavir (EC50s, 0.02 and 0.04 μM, respectively). Moreover, they were active against darunavir (DRV)-resistant variants (EC50 in 0.03 to 0.034 μM range for GRL-04810 and 0.026 to 0.043 μM for GRL-05010), while DRV had EC50s between 0.02 and 0.174 μM. GRL-04810 had a favorable lipophilicity profile as determined with the partition (log P) and distribution (log D) coefficients of -0.14 and -0.29, respectively. The in vitro blood-brain barrier (BBB) permeability assay revealed that GRL-04810 and GRL-05010 may have a greater advantage in terms of crossing the BBB than the currently available PIs, with apparent penetration indexes of 47.8 × 10(-6) and 61.8 × 10(-6) cm/s, respectively. The present data demonstrate that GRL-04810 and GRL-05010 exert efficient activity against a wide spectrum of HIV-1 variants in vitro and suggest that two fluorine atoms added to their bis-THF moieties may well enhance their penetration across the BBB.
Figures




Similar articles
-
GRL-0519, a novel oxatricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor (PI), potently suppresses replication of a wide spectrum of multi-PI-resistant HIV-1 variants in vitro.Antimicrob Agents Chemother. 2013 May;57(5):2036-46. doi: 10.1128/AAC.02189-12. Epub 2013 Feb 12. Antimicrob Agents Chemother. 2013. PMID: 23403426 Free PMC article.
-
A Modified P1 Moiety Enhances In Vitro Antiviral Activity against Various Multidrug-Resistant HIV-1 Variants and In Vitro Central Nervous System Penetration Properties of a Novel Nonpeptidic Protease Inhibitor, GRL-10413.Antimicrob Agents Chemother. 2016 Nov 21;60(12):7046-7059. doi: 10.1128/AAC.01428-16. Print 2016 Dec. Antimicrob Agents Chemother. 2016. PMID: 27620483 Free PMC article.
-
GRL-079, a Novel HIV-1 Protease Inhibitor, Is Extremely Potent against Multidrug-Resistant HIV-1 Variants and Has a High Genetic Barrier against the Emergence of Resistant Variants.Antimicrob Agents Chemother. 2018 Apr 26;62(5):e02060-17. doi: 10.1128/AAC.02060-17. Print 2018 May. Antimicrob Agents Chemother. 2018. PMID: 29463535 Free PMC article.
-
Design of HIV protease inhibitors targeting protein backbone: an effective strategy for combating drug resistance.Acc Chem Res. 2008 Jan;41(1):78-86. doi: 10.1021/ar7001232. Epub 2007 Aug 28. Acc Chem Res. 2008. PMID: 17722874 Review.
-
Darunavir: a review of its use in the management of HIV infection in adults.Drugs. 2009;69(4):477-503. doi: 10.2165/00003495-200969040-00007. Drugs. 2009. PMID: 19323590 Review.
Cited by
-
Management of Antiretroviral Therapy with Boosted Protease Inhibitors-Darunavir/Ritonavir or Darunavir/Cobicistat.Biomedicines. 2021 Mar 18;9(3):313. doi: 10.3390/biomedicines9030313. Biomedicines. 2021. PMID: 33803812 Free PMC article. Review.
-
A novel tricyclic ligand-containing nonpeptidic HIV-1 protease inhibitor, GRL-0739, effectively inhibits the replication of multidrug-resistant HIV-1 variants and has a desirable central nervous system penetration property in vitro.Antimicrob Agents Chemother. 2015 May;59(5):2625-35. doi: 10.1128/AAC.04757-14. Epub 2015 Feb 17. Antimicrob Agents Chemother. 2015. PMID: 25691652 Free PMC article.
-
Novel Central Nervous System (CNS)-Targeting Protease Inhibitors for Drug-Resistant HIV Infection and HIV-Associated CNS Complications.Antimicrob Agents Chemother. 2019 Jun 24;63(7):e00466-19. doi: 10.1128/AAC.00466-19. Print 2019 Jul. Antimicrob Agents Chemother. 2019. PMID: 31061155 Free PMC article.
-
Fluorine Modifications Contribute to Potent Antiviral Activity against Highly Drug-Resistant HIV-1 and Favorable Blood-Brain Barrier Penetration Property of Novel Central Nervous System-Targeting HIV-1 Protease Inhibitors In Vitro.Antimicrob Agents Chemother. 2022 Feb 15;66(2):e0171521. doi: 10.1128/AAC.01715-21. Epub 2022 Jan 3. Antimicrob Agents Chemother. 2022. PMID: 34978889 Free PMC article.
-
Organic carbamates in drug design and medicinal chemistry.J Med Chem. 2015 Apr 9;58(7):2895-940. doi: 10.1021/jm501371s. Epub 2015 Jan 7. J Med Chem. 2015. PMID: 25565044 Free PMC article. Review.
References
-
- Coiras M, Lopez-Huertas M, Perez-Olmeda M, Alcami J. 2009. Understanding HIV-1 latency provides clues for the eradication of long-term reservoirs. Nat. Rev. Microbiol. 7:798–812 - PubMed
-
- De Clercq. 2002. New anti-HIV agents and targets. Med. Res. Rev. 22:531–565 - PubMed
-
- Lewin SR, Murray J, Solomon A, Wightman F, Cameron PU, Purcell DJ, Zaunders JJ, Grey P, Bloch M, Smith D, Cooper DA, Kelleher AD. 2008. Virological determinants of success after structured treatment interruptions of antiretrovirals in acute HIV-1 infection. J. Acquir. Immune Defic. Syndr. 47:140–147 - PubMed
-
- Siliciano JD, Siliciano RF. 2004. A long-term latent reservoir for HIV-1: discovery and clinical implications. J. Antimicrob. Chemother. 54:6–9 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

