Determinants of virulence of influenza A virus
- PMID: 24078062
- PMCID: PMC3969785
- DOI: 10.1007/s10096-013-1984-8
Determinants of virulence of influenza A virus
Abstract
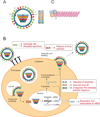
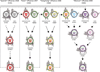
Influenza A viruses cause yearly seasonal epidemics and occasional global pandemics in humans. In the last century, four human influenza A virus pandemics have occurred. Occasionally, influenza A viruses that circulate in other species cross the species barrier and infect humans. Virus reassortment (i.e. mixing of gene segments of multiple viruses) and the accumulation of mutations contribute to the emergence of new influenza A virus variants. Fortunately, most of these variants do not have the ability to spread among humans and subsequently cause a pandemic. In this review, we focus on the threat of animal influenza A viruses which have shown the ability to infect humans. In addition, genetic factors which could alter the virulence of influenza A viruses are discussed. The identification and characterisation of these factors may provide insights into genetic traits which change virulence and help us to understand which genetic determinants are of importance for the pandemic potential of animal influenza A viruses.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures
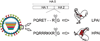
Similar articles
-
Virulence determinants of pandemic influenza viruses.J Clin Invest. 2011 Jan;121(1):6-13. doi: 10.1172/JCI44947. Epub 2011 Jan 4. J Clin Invest. 2011. PMID: 21206092 Free PMC article. Review.
-
Virulence and genetic compatibility of polymerase reassortant viruses derived from the pandemic (H1N1) 2009 influenza virus and circulating influenza A viruses.J Virol. 2011 Jul;85(13):6275-86. doi: 10.1128/JVI.02125-10. Epub 2011 Apr 20. J Virol. 2011. PMID: 21507962 Free PMC article.
-
Avian influenza virus transmission to mammals.Curr Top Microbiol Immunol. 2014;385:137-55. doi: 10.1007/82_2014_387. Curr Top Microbiol Immunol. 2014. PMID: 25048542 Review.
-
[Influenza pandemic: hypotheses and facts].Zh Mikrobiol Epidemiol Immunobiol. 2008 Sep-Oct;(5):109-18. Zh Mikrobiol Epidemiol Immunobiol. 2008. PMID: 19004295 Review. Russian.
-
Recent zoonoses caused by influenza A viruses.Rev Sci Tech. 2000 Apr;19(1):197-225. doi: 10.20506/rst.19.1.1220. Rev Sci Tech. 2000. PMID: 11189716 Review.
Cited by
-
Scoring amino acid mutation to predict pandemic risk of avian influenza virus.BMC Bioinformatics. 2019 Jun 10;20(Suppl 8):288. doi: 10.1186/s12859-019-2770-0. BMC Bioinformatics. 2019. PMID: 31182019 Free PMC article.
-
Detection of reassortant influenza B strains from 2004 to 2015 seasons in Barcelona (Catalonia, Spain) by whole genome sequencing.Virus Res. 2023 Jun;330:199089. doi: 10.1016/j.virusres.2023.199089. Epub 2023 Apr 5. Virus Res. 2023. PMID: 37011863 Free PMC article.
-
Viral-Host Interactome Analysis Reveals Chicken STAU2 Interacts With Non-structural Protein 1 and Promotes the Replication of H5N1 Avian Influenza Virus.Front Immunol. 2021 Apr 21;12:590679. doi: 10.3389/fimmu.2021.590679. eCollection 2021. Front Immunol. 2021. PMID: 33968009 Free PMC article.
-
Comparison of PB1-F2 Proximity Interactomes Reveals Functional Differences between a Human and an Avian Influenza Virus.Viruses. 2023 Jan 24;15(2):328. doi: 10.3390/v15020328. Viruses. 2023. PMID: 36851542 Free PMC article.
-
Inflammation and Pneumonia: Why Are Some More Susceptible than Others?Clin Chest Med. 2018 Dec;39(4):669-676. doi: 10.1016/j.ccm.2018.07.002. Clin Chest Med. 2018. PMID: 30390740 Free PMC article. Review.
References
-
- Palese P, Shaw ML. Fields Virology Lippincott. Philadelphia, USA: Williams & Wilkins; 2007. pp. p 1647–p 1690.
-
- Wise HM, Hutchinson EC, Jagger BW, Stuart AD, Kang ZH, Robb N, Schwartzman LM, Kash JC, Fodor E, Firth AE, Gog JR, Taubenberger JK, Digard P. Identification of a novel splice variant form of the influenza a virus m2 ion channel with an antigenically distinct ectodomain. PLoS Pathog. 2012;8(11):e1002998. - PMC - PubMed
-
- Jagger BW, Wise HM, Kash JC, Walters KA, Wills NM, Xiao YL, Dunfee RL, Schwartzman LM, Ozinsky A, Bell GL, Dalton RM, Lo A, Efstathiou S, Atkins JF, Firth AE, Taubenberger JK, Digard P. An overlapping protein-coding region in influenza A virus segment 3 modulates the host response. Science. 2012;337(6091):199–204. - PMC - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

