Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models
- PMID: 24067968
- PMCID: PMC3838248
- DOI: 10.1128/JVI.02420-13
Polycomb repressive complex 2 silences human cytomegalovirus transcription in quiescent infection models
Abstract
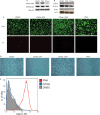
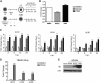
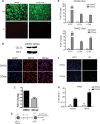
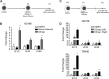
Chromatin-based regulation of herpesviral transcriptional programs is increasingly appreciated as a mechanism for modulating infection outcomes. Transcriptionally permissive euchromatin predominates during lytic infection, whereas heterochromatin domains refractory to transcription are enriched at lytic genes during latency. Reversibly silenced facultative heterochromatin domains are often enriched for histone H3 trimethylated on lysine 27 (H3K27me3), a modification catalyzed by Polycomb repressive complex 2 (PRC2). The requirement for PRC2 in suppressing the human cytomegalovirus (HCMV) lytic transcriptional program during latency has not been thoroughly evaluated. Therefore, we disrupted PRC2 activity in the highly tractable THP1 and NT2D1 quiescent-infection models by treating cells with small-molecule inhibitors of PRC2 activity. Compared to control cells, disruption of PRC2 in HCMV-infected THP1 or NT2D1 cells resulted in significant increases in viral transcript levels and the detection of viral protein. Using chromatin immunoprecipitation, we demonstrated that enrichment of H3K27me3, deposited by PRC2, correlates inversely with lytic transcriptional output, suggesting that PRC2 catalytic activity at viral chromatin directly represses lytic transcription. Together, our data suggest that PRC2-mediated repression of viral transcription is a key step in the establishment and maintenance of HCMV latency.
Figures
Similar articles
-
A Noncanonical Function of Polycomb Repressive Complexes Promotes Human Cytomegalovirus Lytic DNA Replication and Serves as a Novel Cellular Target for Antiviral Intervention.J Virol. 2019 Apr 17;93(9):e02143-18. doi: 10.1128/JVI.02143-18. Print 2019 May 1. J Virol. 2019. PMID: 30814291 Free PMC article.
-
Kinetics of facultative heterochromatin and polycomb group protein association with the herpes simplex viral genome during establishment of latent infection.mBio. 2013 Jan 15;4(1):e00590-12. doi: 10.1128/mBio.00590-12. mBio. 2013. PMID: 23322639 Free PMC article.
-
IMITATION SWITCH is required for normal chromatin structure and gene repression in PRC2 target domains.Proc Natl Acad Sci U S A. 2021 Jan 26;118(4):e2010003118. doi: 10.1073/pnas.2010003118. Proc Natl Acad Sci U S A. 2021. PMID: 33468665 Free PMC article.
-
Chromatin-mediated regulation of cytomegalovirus gene expression.Virus Res. 2011 May;157(2):134-43. doi: 10.1016/j.virusres.2010.09.019. Epub 2010 Sep 25. Virus Res. 2011. PMID: 20875471 Free PMC article. Review.
-
Chromatin structure regulates human cytomegalovirus gene expression during latency, reactivation and lytic infection.Biochim Biophys Acta. 2010 Mar-Apr;1799(3-4):286-95. doi: 10.1016/j.bbagrm.2009.08.001. Epub 2009 Aug 12. Biochim Biophys Acta. 2010. PMID: 19682613 Review.
Cited by
-
A BMPR2/YY1 Signaling Axis Is Required for Human Cytomegalovirus Latency in Undifferentiated Myeloid Cells.mBio. 2021 Jun 29;12(3):e0022721. doi: 10.1128/mBio.00227-21. Epub 2021 Jun 1. mBio. 2021. PMID: 34061599 Free PMC article.
-
Cellular defense against latent colonization foiled by human cytomegalovirus UL138 protein.Sci Adv. 2015 Nov 27;1(10):e1501164. doi: 10.1126/sciadv.1501164. eCollection 2015 Nov. Sci Adv. 2015. PMID: 26702450 Free PMC article.
-
Epigenetics and Genetics of Viral Latency.Cell Host Microbe. 2016 May 11;19(5):619-28. doi: 10.1016/j.chom.2016.04.008. Cell Host Microbe. 2016. PMID: 27173930 Free PMC article. Review.
-
Targeting Host Cellular Factors as a Strategy of Therapeutic Intervention for Herpesvirus Infections.Front Cell Infect Microbiol. 2021 Mar 19;11:603309. doi: 10.3389/fcimb.2021.603309. eCollection 2021. Front Cell Infect Microbiol. 2021. PMID: 33816328 Free PMC article. Review.
-
Human cytomegalovirus reactivation from latency: validation of a "switch" model in vitro.Virol J. 2016 Oct 22;13(1):179. doi: 10.1186/s12985-016-0634-z. Virol J. 2016. PMID: 27770817 Free PMC article.
References
-
- Mocarski ES, Shenk T, Pass RF. 2007. Cytomegaloviruses, p 2701–2772 In Knipe DM, Howley PM, Griffin DE, Lamb RA, Martin MA, Roizman B, Straus SE. (ed), Fields virology, 5th ed, vol 2 Lippincott Williams & Wilkins, Philadelphia, PA
-
- Sinclair J. 2008. Human cytomegalovirus: latency and reactivation in the myeloid lineage. J. Clin. Virol. 41:180–185 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

