Comprehensive mapping and analysis of Kaposi's sarcoma-associated herpesvirus 3' UTRs identify differential posttranscriptional control of gene expression in lytic versus latent infection
- PMID: 24067953
- PMCID: PMC3838127
- DOI: 10.1128/JVI.02374-13
Comprehensive mapping and analysis of Kaposi's sarcoma-associated herpesvirus 3' UTRs identify differential posttranscriptional control of gene expression in lytic versus latent infection
Abstract
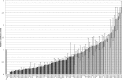
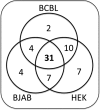
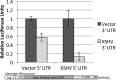
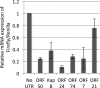
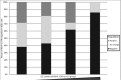
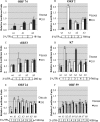
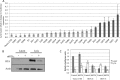
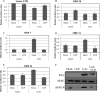
3' untranslated regions (UTRs) are known to play an important role in posttranscriptional regulation of gene expression. Here we map the 3' UTRs of Kaposi's sarcoma-associated herpesvirus (KSHV) using next-generation RNA sequencing, 3' rapid amplification of cDNA ends (RACE), and tiled microarray analyses. Chimeric reporters containing the KSHV 3' UTRs show a general trend toward reduced gene expression under conditions of latent infection. Those 3' UTRs with a higher GC content are more likely to be associated with reduced gene expression. KSHV transcripts display an extensive use of shared polyadenylation sites allowing for partially overlapping 3' UTRs and regulatory activities. In addition, a subset of KSHV 3' UTRs is sufficient to convey increased gene expression under conditions of lytic infection. These results suggest a role for viral 3' UTRs in contributing to differential gene expression during latent versus lytic infection.
Figures

Similar articles
-
Identification of the physiological gene targets of the essential lytic replicative Kaposi's sarcoma-associated herpesvirus ORF57 protein.J Virol. 2015 Feb;89(3):1688-702. doi: 10.1128/JVI.02663-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410858 Free PMC article.
-
Transcriptome analysis of Kaposi's sarcoma-associated herpesvirus during de novo primary infection of human B and endothelial cells.J Virol. 2015 Mar;89(6):3093-111. doi: 10.1128/JVI.02507-14. Epub 2014 Dec 31. J Virol. 2015. PMID: 25552714 Free PMC article.
-
Kaposi's sarcoma-associated herpesvirus-encoded latency-associated nuclear antigen inhibits lytic replication by targeting Rta: a potential mechanism for virus-mediated control of latency.J Virol. 2004 Jun;78(12):6585-94. doi: 10.1128/JVI.78.12.6585-6594.2004. J Virol. 2004. PMID: 15163750 Free PMC article.
-
Towards Better Understanding of KSHV Life Cycle: from Transcription and Posttranscriptional Regulations to Pathogenesis.Virol Sin. 2019 Apr;34(2):135-161. doi: 10.1007/s12250-019-00114-3. Epub 2019 Apr 25. Virol Sin. 2019. PMID: 31025296 Free PMC article. Review.
-
Epigenetic regulation of Kaposi's sarcoma-associated herpesvirus replication.Semin Cancer Biol. 2009 Jun;19(3):153-7. doi: 10.1016/j.semcancer.2009.02.010. Epub 2009 Feb 21. Semin Cancer Biol. 2009. PMID: 19429478 Free PMC article. Review.
Cited by
-
KSHV 3.0: a state-of-the-art annotation of the Kaposi's sarcoma-associated herpesvirus transcriptome using cross-platform sequencing.mSystems. 2024 Feb 20;9(2):e0100723. doi: 10.1128/msystems.01007-23. Epub 2024 Jan 11. mSystems. 2024. PMID: 38206015 Free PMC article.
-
KSHV 2.0: a comprehensive annotation of the Kaposi's sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features.PLoS Pathog. 2014 Jan;10(1):e1003847. doi: 10.1371/journal.ppat.1003847. Epub 2014 Jan 16. PLoS Pathog. 2014. PMID: 24453964 Free PMC article.
-
MicroRNAs Expressed during Viral Infection: Biomarker Potential and Therapeutic Considerations.Biomark Insights. 2016 Jan 18;10(Suppl 4):25-52. doi: 10.4137/BMI.S29512. eCollection 2015. Biomark Insights. 2016. PMID: 26819546 Free PMC article. Review.
-
Identification of the physiological gene targets of the essential lytic replicative Kaposi's sarcoma-associated herpesvirus ORF57 protein.J Virol. 2015 Feb;89(3):1688-702. doi: 10.1128/JVI.02663-14. Epub 2014 Nov 19. J Virol. 2015. PMID: 25410858 Free PMC article.
-
Today's Kaposi sarcoma is not the same as it was 40 years ago, or is it?J Med Virol. 2023 May;95(5):e28773. doi: 10.1002/jmv.28773. J Med Virol. 2023. PMID: 37212317 Free PMC article. Review.
References
-
- Elkon R, Ugalde AP, Agami R. 2013. Alternative cleavage and polyadenylation: extent, regulation and function. Nat. Rev. Genet. 14:496–506 - PubMed
-
- Calin GA, Liu CG, Sevignani C, Ferracin M, Felli N, Dumitru CD, Shimizu M, Cimmino A, Zupo S, Dono M, Dell'Aquila ML, Alder H, Rassenti L, Kipps TJ, Bullrich F, Negrini M, Croce CM. 2004. MicroRNA profiling reveals distinct signatures in B cell chronic lymphocytic leukemias. Proc. Natl. Acad. Sci. U. S. A. 101:11755–11760 - PMC - PubMed
-
- Shaw G, Kamen R. 1986. A conserved AU sequence from the 3′ untranslated region of GM-CSF mRNA mediates selective mRNA degradation. Cell 46:659–667 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

