Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency
- PMID: 24055366
- PMCID: PMC3790458
- DOI: 10.1016/j.cell.2013.08.032
Mitochondrial cristae shape determines respiratory chain supercomplexes assembly and respiratory efficiency
Abstract
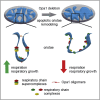
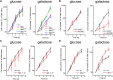
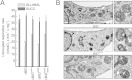
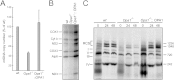
Respiratory chain complexes assemble into functional quaternary structures called supercomplexes (RCS) within the folds of the inner mitochondrial membrane, or cristae. Here, we investigate the relationship between respiratory function and mitochondrial ultrastructure and provide evidence that cristae shape determines the assembly and stability of RCS and hence mitochondrial respiratory efficiency. Genetic and apoptotic manipulations of cristae structure affect assembly and activity of RCS in vitro and in vivo, independently of changes to mitochondrial protein synthesis or apoptotic outer mitochondrial membrane permeabilization. We demonstrate that, accordingly, the efficiency of mitochondria-dependent cell growth depends on cristae shape. Thus, RCS assembly emerges as a link between membrane morphology and function.
Copyright © 2013 The Authors. Published by Elsevier Inc. All rights reserved.
Figures







Comment in
-
Mitochondria: organization of respiratory chain complexes becomes cristae-lized.Curr Biol. 2013 Nov 4;23(21):R969-71. doi: 10.1016/j.cub.2013.09.035. Curr Biol. 2013. PMID: 24200328
Similar articles
-
Who and how in the regulation of mitochondrial cristae shape and function.Biochem Biophys Res Commun. 2018 May 27;500(1):94-101. doi: 10.1016/j.bbrc.2017.04.088. Epub 2017 Apr 21. Biochem Biophys Res Commun. 2018. PMID: 28438601 Review.
-
OPA1 regulates respiratory supercomplexes assembly: The role of mitochondrial swelling.Mitochondrion. 2020 Mar;51:30-39. doi: 10.1016/j.mito.2019.11.006. Epub 2019 Dec 20. Mitochondrion. 2020. PMID: 31870826 Free PMC article.
-
Bid maintains mitochondrial cristae structure and function and protects against cardiac disease in an integrative genomics study.Elife. 2018 Oct 3;7:e40907. doi: 10.7554/eLife.40907. Elife. 2018. PMID: 30281024 Free PMC article.
-
Opa1-mediated cristae opening is Bax/Bak and BH3 dependent, required for apoptosis, and independent of Bak oligomerization.Mol Cell. 2008 Aug 22;31(4):557-569. doi: 10.1016/j.molcel.2008.07.010. Epub 2008 Aug 7. Mol Cell. 2008. PMID: 18691924 Free PMC article.
-
Cardiolipin and mitochondrial cristae organization.Biochim Biophys Acta Biomembr. 2017 Jun;1859(6):1156-1163. doi: 10.1016/j.bbamem.2017.03.013. Epub 2017 Mar 20. Biochim Biophys Acta Biomembr. 2017. PMID: 28336315 Free PMC article. Review.
Cited by
-
The potential mechanism of mitochondrial homeostasis in postoperative neurocognitive disorders: an in-depth review.Ann Med. 2024 Dec;56(1):2411012. doi: 10.1080/07853890.2024.2411012. Epub 2024 Oct 25. Ann Med. 2024. PMID: 39450938 Free PMC article. Review.
-
Mitochondrial Quality Control Orchestrates the Symphony of B Cells and Plays Critical Roles in B Cell-Related Diseases.J Immunol Res. 2024 Oct 17;2024:5577506. doi: 10.1155/2024/5577506. eCollection 2024. J Immunol Res. 2024. PMID: 39449998 Free PMC article. Review.
-
Calorie restriction increases insulin sensitivity to promote beta cell homeostasis and longevity in mice.Nat Commun. 2024 Oct 21;15(1):9063. doi: 10.1038/s41467-024-53127-2. Nat Commun. 2024. PMID: 39433757 Free PMC article.
-
Targeting DNM1L/DRP1-FIS1 axis inhibits high-grade glioma progression by impeding mitochondrial respiratory cristae remodeling.J Exp Clin Cancer Res. 2024 Sep 30;43(1):273. doi: 10.1186/s13046-024-03194-6. J Exp Clin Cancer Res. 2024. PMID: 39350223 Free PMC article.
-
Insights into Transient Dimerisation of Carnitine/Acylcarnitine Carrier (SLC25A20) from Sarkosyl/PAGE, Cross-Linking Reagents, and Comparative Modelling Analysis.Biomolecules. 2024 Sep 14;14(9):1158. doi: 10.3390/biom14091158. Biomolecules. 2024. PMID: 39334924 Free PMC article.
References
-
- Acín-Pérez R., Fernández-Silva P., Peleato M.L., Pérez-Martos A., Enriquez J.A. Respiratory active mitochondrial supercomplexes. Mol. Cell. 2008;32:529–539. - PubMed
-
- Alexander C., Votruba M., Pesch U.E., Thiselton D.L., Mayer S., Moore A., Rodriguez M., Kellner U., Leo-Kottler B., Auburger G. OPA1, encoding a dynamin-related GTPase, is mutated in autosomal dominant optic atrophy linked to chromosome 3q28. Nat. Genet. 2000;26:211–215. - PubMed
-
- Campanella M., Casswell E., Chong S., Farah Z., Wieckowski M.R., Abramov A.Y., Tinker A., Duchen M.R. Regulation of mitochondrial structure and function by the F1Fo-ATPase inhibitor protein, IF1. Cell Metab. 2008;8:13–25. - PubMed
Supplemental References
-
- Dimmer K.S., Navoni F., Casarin A., Trevisson E., Endele S., Winterpacht A., Salviati L., Scorrano L. LETM1, deleted in Wolf-Hirschhorn syndrome is required for normal mitochondrial morphology and cellular viability. Hum. Mol. Genet. 2008;17:201–214. - PubMed
-
- Frezza C., Cipolat S., Scorrano L. Organelle isolation: functional mitochondria from mouse liver, muscle and cultured fibroblasts. Nat. Protoc. 2007;2:287–295. - PubMed
-
- Spinazzi M., Casarin A., Pertegato V., Salviati L., Angelini C. Assessment of mitochondrial respiratory chain enzymatic activities on tissues and cultured cells. Nat. Protoc. 2012;7:1235–1246. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

