Expression of the placental transcriptome in maternal nutrient reduction in baboons is dependent on fetal sex
- PMID: 24047701
- PMCID: PMC3796342
- DOI: 10.3945/jn.112.172148
Expression of the placental transcriptome in maternal nutrient reduction in baboons is dependent on fetal sex
Abstract
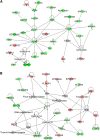
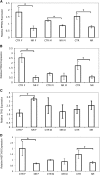
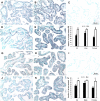
Maternal undernutrition increases the risk of perinatal complications and predisposes offspring to obesity, diabetes, and cardiovascular disease later in life. Emerging evidence suggests that changes in placental function play a role in linking altered maternal nutrition in pregnancy to the subsequent development of adult disease. The susceptibility for disease in response to an adverse intrauterine environment differs distinctly between boys and girls, with girls typically having better outcomes. Here, we tested the hypothesis that regulation of the placental transcriptome by maternal nutrient reduction (NR) is dependent on fetal sex. We used a nonhuman primate model of NR in which maternal global food intake was reduced by 30% in baboons starting at gestational day (GD) 30. At GD 165 (term = GD 183), placental genome expression profiling of 6 control (n = 3 females, 3 males) and 6 nutrient restricted (n = 3 females, 3 males) fetuses was carried out followed by bioinformatic analysis. Surprisingly, there was no coordinated placental molecular response to decreased nutrient availability when analyzing the data without accounting for fetal sex. In contrast, female placentas exhibited a highly coordinated response that included upregulation of genes in networks, pathways, and functional groups related to programmed cell death and downregulation of genes in networks, pathways, and functional groups associated with cell proliferation. These changes were not apparent in the male placentas. Our data support the concept that female placentas initiate complex adaptive responses to an adverse intrauterine environment, which may contribute to increased survival and better pregnancy outcomes in girls.
Conflict of interest statement
Author disclosures: L. A. Cox, C. Li, J. P. Glenn, K. Lange, K. D. Spradling, P. W. Nathanielsz, and T. Jansson, no conflicts of interest.
Figures
Similar articles
-
Effect of 30 per cent maternal nutrient restriction from 0.16 to 0.5 gestation on fetal baboon kidney gene expression.J Physiol. 2006 Apr 1;572(Pt 1):67-85. doi: 10.1113/jphysiol.2006.106872. Epub 2006 Mar 2. J Physiol. 2006. PMID: 16513668 Free PMC article.
-
Reduced placental amino acid transport in response to maternal nutrient restriction in the baboon.Am J Physiol Regul Integr Comp Physiol. 2015 Oct;309(7):R740-6. doi: 10.1152/ajpregu.00161.2015. Epub 2015 Aug 5. Am J Physiol Regul Integr Comp Physiol. 2015. PMID: 26246504 Free PMC article.
-
Down-Regulation of Placental Transport of Amino Acids Precedes the Development of Intrauterine Growth Restriction in Maternal Nutrient Restricted Baboons.Biol Reprod. 2016 Nov;95(5):98. doi: 10.1095/biolreprod.116.141085. Epub 2016 Sep 7. Biol Reprod. 2016. PMID: 27605346 Free PMC article.
-
Effects of Maternal Obesity On Placental Phenotype.Curr Vasc Pharmacol. 2021;19(2):113-131. doi: 10.2174/1570161118666200513115316. Curr Vasc Pharmacol. 2021. PMID: 32400334 Review.
-
Human Placental Adaptive Changes in Response to Maternal Obesity: Sex Specificities.Int J Mol Sci. 2023 Jun 5;24(11):9770. doi: 10.3390/ijms24119770. Int J Mol Sci. 2023. PMID: 37298720 Free PMC article. Review.
Cited by
-
The baboon (Papio sp.) as a model for female reproduction studies.Contraception. 2015 Aug;92(2):120-3. doi: 10.1016/j.contraception.2015.06.007. Epub 2015 Jun 12. Contraception. 2015. PMID: 26072741 Free PMC article. Review.
-
Modulation of Placental Gene Expression in Small-for-Gestational-Age Infants.Genes (Basel). 2020 Jan 10;11(1):80. doi: 10.3390/genes11010080. Genes (Basel). 2020. PMID: 31936801 Free PMC article. Review.
-
Strength of nonhuman primate studies of developmental programming: review of sample sizes, challenges, and steps for future work.J Dev Orig Health Dis. 2020 Jun;11(3):297-306. doi: 10.1017/S2040174419000539. Epub 2019 Sep 30. J Dev Orig Health Dis. 2020. PMID: 31566171 Free PMC article. Review.
-
Effect of moderate, 30 percent global maternal nutrient reduction on fetal and postnatal baboon phenotype.J Med Primatol. 2017 Dec;46(6):293-303. doi: 10.1111/jmp.12290. Epub 2017 Jul 26. J Med Primatol. 2017. PMID: 28744866 Free PMC article.
-
Maternal dyslipidemia during early pregnancy and epigenetic ageing of the placenta.Epigenetics. 2019 Oct;14(10):1030-1039. doi: 10.1080/15592294.2019.1629234. Epub 2019 Jun 14. Epigenetics. 2019. PMID: 31179827 Free PMC article.
References
-
- Gluckman PD, Hanson MA. Living with the past: evolution, development, and patterns of disease. Science. 2004;305:1733–6. - PubMed
-
- Barker DJP. The developmental origins of insulin resistance. Horm Res. 2005;64 Suppl 3:2–7. - PubMed
-
- Jansson T, Powell TL. Role of the placenta in fetal programming: Underlying mechanisms and potential interventional approaches. Clin Sci (Lond). 2007;113:1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

