Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping
- PMID: 24009888
- PMCID: PMC3760630
- DOI: 10.3402/jev.v2i0.20920
Extracellular Vesicle (EV) Array: microarray capturing of exosomes and other extracellular vesicles for multiplexed phenotyping
Abstract
Background: Exosomes are one of the several types of cell-derived vesicles with a diameter of 30-100 nm. These extracellular vesicles are recognized as potential markers of human diseases such as cancer. However, their use in diagnostic tests requires an objective and high-throughput method to define their phenotype and determine their concentration in biological fluids. To identify circulating as well as cell culture-derived vesicles, the current standard is immunoblotting or a flow cytometrical analysis for specific proteins, both of which requires large amounts of purified vesicles.
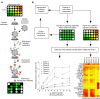
Methods: Based on the technology of protein microarray, we hereby present a highly sensitive Extracellular Vesicle (EV) Array capable of detecting and phenotyping exosomes and other extracellular vesicles from unpurified starting material in a high-throughput manner. To only detect the exosomes captured on the EV Array, a cocktail of antibodies against the tetraspanins CD9, CD63 and CD81 was used. These antibodies were selected to ensure that all exosomes captured are detected, and concomitantly excluding the detection of other types of microvesicles.
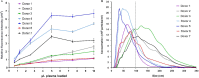
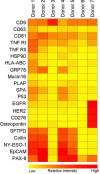
Results: The limit of detection (LOD) was determined on exosomes derived from the colon cancer cell line LS180. It clarified that supernatant from only approximately 10(4) cells was needed to obtain signals or that only 2.5×10(4) exosomes were required for each microarray spot (~1 nL). Phenotyping was performed on plasma (1-10 µL) from 7 healthy donors, which were applied to the EV Array with a panel of antibodies against 21 different cellular surface antigens and cancer antigens. For each donor, there was considerable heterogeneity in the expression levels of individual markers. The protein profiles of the exosomes (defined as positive for CD9, CD63 and CD81) revealed that only the expression level of CD9 and CD81 was approximately equal in the 7 donors. This implies questioning the use of CD63 as a standard exosomal marker since the expression level of this tetraspanin was considerably lower.
Keywords: EV Array; antigenic capturing; exosomes; extracellular vesicles; nanoparticle tracking analysis; phenotyping; plasma; protein microarray.
Figures

Similar articles
-
Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma.J Extracell Vesicles. 2015 Mar 2;4:26659. doi: 10.3402/jev.v4.26659. eCollection 2015. J Extracell Vesicles. 2015. PMID: 25735706 Free PMC article.
-
Nanoscale flow cytometry to distinguish subpopulations of prostate extracellular vesicles in patient plasma.Prostate. 2019 May;79(6):592-603. doi: 10.1002/pros.23764. Epub 2019 Jan 24. Prostate. 2019. PMID: 30680751
-
Multiplexed Phenotyping of Small Extracellular Vesicles Using Protein Microarray (EV Array).Methods Mol Biol. 2017;1545:117-127. doi: 10.1007/978-1-4939-6728-5_8. Methods Mol Biol. 2017. PMID: 27943210
-
Plasma neuronal exosomes serve as biomarkers of cognitive impairment in HIV infection and Alzheimer's disease.J Neurovirol. 2019 Oct;25(5):702-709. doi: 10.1007/s13365-018-0695-4. Epub 2019 Jan 4. J Neurovirol. 2019. PMID: 30610738 Free PMC article. Review.
-
Extracellular Vesicles: Recent Developments in Technology and Perspectives for Cancer Liquid Biopsy.Recent Results Cancer Res. 2020;215:319-344. doi: 10.1007/978-3-030-26439-0_17. Recent Results Cancer Res. 2020. PMID: 31605237 Review.
Cited by
-
Exosomal proteins as potential diagnostic markers in advanced non-small cell lung carcinoma.J Extracell Vesicles. 2015 Mar 2;4:26659. doi: 10.3402/jev.v4.26659. eCollection 2015. J Extracell Vesicles. 2015. PMID: 25735706 Free PMC article.
-
Potentials and capabilities of the Extracellular Vesicle (EV) Array.J Extracell Vesicles. 2015 Apr 8;4:26048. doi: 10.3402/jev.v4.26048. eCollection 2015. J Extracell Vesicles. 2015. PMID: 25862471 Free PMC article.
-
Microfluidic Exosome Analysis toward Liquid Biopsy for Cancer.J Lab Autom. 2016 Aug;21(4):599-608. doi: 10.1177/2211068216651035. Epub 2016 May 23. J Lab Autom. 2016. PMID: 27215792 Free PMC article. Review.
-
Emerging micro-nanotechnologies for extracellular vesicles in immuno-oncology: from target specific isolations to immunomodulation.Lab Chip. 2022 Sep 13;22(18):3314-3339. doi: 10.1039/d2lc00232a. Lab Chip. 2022. PMID: 35980234 Free PMC article. Review.
-
Direct isolation of small extracellular vesicles from human blood using viscoelastic microfluidics.Sci Adv. 2023 Oct 6;9(40):eadi5296. doi: 10.1126/sciadv.adi5296. Epub 2023 Oct 6. Sci Adv. 2023. PMID: 37801500 Free PMC article.
References
-
- Lee TH, D'Asti E, Magnus N, Al-Nedawi K, Meehan B, Rak J. Microvesicles as mediators of intercellular communication in cancer – the emerging science of cellular “debris”. Semin Immunopathol. 2011;33:455–67. - PubMed
-
- Simons M, Raposo G. Exosomes-vesicular carriers for intercellular communication. Curr Opin Cell Biol. 2009;21:575–81. - PubMed
-
- Théry C, Ostrowski M, Segura E. Membrane vesicles as conveyors of immune responses. Nature reviews. Immunology. 2009;9:581–93. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

