Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens
- PMID: 23995682
- PMCID: PMC3825626
- DOI: 10.1038/nature12503
Microbiota-liberated host sugars facilitate post-antibiotic expansion of enteric pathogens
Abstract
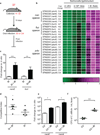
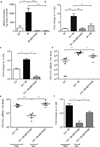
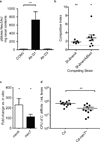
The human intestine, colonized by a dense community of resident microbes, is a frequent target of bacterial pathogens. Undisturbed, this intestinal microbiota provides protection from bacterial infections. Conversely, disruption of the microbiota with oral antibiotics often precedes the emergence of several enteric pathogens. How pathogens capitalize upon the failure of microbiota-afforded protection is largely unknown. Here we show that two antibiotic-associated pathogens, Salmonella enterica serovar Typhimurium (S. typhimurium) and Clostridium difficile, use a common strategy of catabolizing microbiota-liberated mucosal carbohydrates during their expansion within the gut. S. typhimurium accesses fucose and sialic acid within the lumen of the gut in a microbiota-dependent manner, and genetic ablation of the respective catabolic pathways reduces its competitiveness in vivo. Similarly, C. difficile expansion is aided by microbiota-induced elevation of sialic acid levels in vivo. Colonization of gnotobiotic mice with a sialidase-deficient mutant of Bacteroides thetaiotaomicron, a model gut symbiont, reduces free sialic acid levels resulting in C. difficile downregulating its sialic acid catabolic pathway and exhibiting impaired expansion. These effects are reversed by exogenous dietary administration of free sialic acid. Furthermore, antibiotic treatment of conventional mice induces a spike in free sialic acid and mutants of both Salmonella and C. difficile that are unable to catabolize sialic acid exhibit impaired expansion. These data show that antibiotic-induced disruption of the resident microbiota and subsequent alteration in mucosal carbohydrate availability are exploited by these two distantly related enteric pathogens in a similar manner. This insight suggests new therapeutic approaches for preventing diseases caused by antibiotic-associated pathogens.
Figures
Comment in
-
Finding a sugary foothold: how antibiotics pave the way for enteric pathogens.Cell Host Microbe. 2013 Sep 11;14(3):225-7. doi: 10.1016/j.chom.2013.08.008. Cell Host Microbe. 2013. PMID: 24034607
-
An antibiotic-altered microbiota provides fuel for the enteric foe.Cell Res. 2014 Jan;24(1):5-6. doi: 10.1038/cr.2013.142. Epub 2013 Oct 29. Cell Res. 2014. PMID: 24165893 Free PMC article.
-
How enteric pathogens know they hit the sweet spot.Future Microbiol. 2014;9(1):13-6. doi: 10.2217/fmb.13.141. Future Microbiol. 2014. PMID: 24328376 Free PMC article.
Similar articles
-
An antibiotic-altered microbiota provides fuel for the enteric foe.Cell Res. 2014 Jan;24(1):5-6. doi: 10.1038/cr.2013.142. Epub 2013 Oct 29. Cell Res. 2014. PMID: 24165893 Free PMC article.
-
How enteric pathogens know they hit the sweet spot.Future Microbiol. 2014;9(1):13-6. doi: 10.2217/fmb.13.141. Future Microbiol. 2014. PMID: 24328376 Free PMC article.
-
Finding a sugary foothold: how antibiotics pave the way for enteric pathogens.Cell Host Microbe. 2013 Sep 11;14(3):225-7. doi: 10.1016/j.chom.2013.08.008. Cell Host Microbe. 2013. PMID: 24034607
-
Role of the intestinal microbiota in resistance to colonization by Clostridium difficile.Gastroenterology. 2014 May;146(6):1547-53. doi: 10.1053/j.gastro.2014.01.059. Epub 2014 Feb 4. Gastroenterology. 2014. PMID: 24503131 Free PMC article. Review.
-
Establishing causality in Salmonella-microbiota-host interaction: The use of gnotobiotic mouse models and synthetic microbial communities.Int J Med Microbiol. 2021 Apr;311(3):151484. doi: 10.1016/j.ijmm.2021.151484. Epub 2021 Mar 2. Int J Med Microbiol. 2021. PMID: 33756190 Review.
Cited by
-
C. difficile exploits a host metabolite produced during toxin-mediated disease.Nature. 2021 May;593(7858):261-265. doi: 10.1038/s41586-021-03502-6. Epub 2021 Apr 28. Nature. 2021. PMID: 33911281 Free PMC article.
-
Metabolite dependence of antibiotic susceptibility in a gut microbe.mSphere. 2024 Oct 29;9(10):e0060024. doi: 10.1128/msphere.00600-24. Epub 2024 Sep 19. mSphere. 2024. PMID: 39297638 Free PMC article.
-
Mucin glycan foraging in the human gut microbiome.Front Genet. 2015 Mar 19;6:81. doi: 10.3389/fgene.2015.00081. eCollection 2015. Front Genet. 2015. PMID: 25852737 Free PMC article. Review.
-
Intestinal epithelial glycosylation in homeostasis and gut microbiota interactions in IBD.Nat Rev Gastroenterol Hepatol. 2020 Oct;17(10):597-617. doi: 10.1038/s41575-020-0331-7. Epub 2020 Jul 24. Nat Rev Gastroenterol Hepatol. 2020. PMID: 32710014 Free PMC article. Review.
-
Host-Derived Sialic Acids Are an Important Nutrient Source Required for Optimal Bacterial Fitness In Vivo.mBio. 2016 Apr 12;7(2):e02237-15. doi: 10.1128/mBio.02237-15. mBio. 2016. PMID: 27073099 Free PMC article.
References
-
- Doorduyn Y, Van Den Brandhof WE, Van Duynhoven YT, Wannet WJ, Van Pelt W. Risk factors for Salmonella Enteritidis and Typhimurium (DT104 and non-DT104) infections in The Netherlands: predominant roles for raw eggs in Enteritidis and sandboxes in Typhimurium infections. Epidemiology and infection. 2006;134:617–626. - PMC - PubMed
-
- Pavia AT, et al. Epidemiologic evidence that prior antimicrobial exposure decreases resistance to infection by antimicrobial-sensitive Salmonella. The Journal of infectious diseases. 1990;161:255–260. - PubMed
-
- Pepin J, et al. Emergence of fluoroquinolones as the predominant risk factor for Clostridium difficile-associated diarrhea: a cohort study during an epidemic in Quebec. Clin Infect Dis. 2005;41:1254–1260. - PubMed
-
- Kelly CP, Pothoulakis C, LaMont JT. Clostridium difficile colitis. N Engl J Med. 1994;330:257–262. - PubMed
-
- Backhed F, Ley RE, Sonnenburg JL, Peterson DA, Gordon JI. Host-bacterial mutualism in the human intestine. Science (New York, N.Y. 2005;307:1915–1920. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

