Topoisomerases facilitate transcription of long genes linked to autism
- PMID: 23995680
- PMCID: PMC3767287
- DOI: 10.1038/nature12504
Topoisomerases facilitate transcription of long genes linked to autism
Abstract
Topoisomerases are expressed throughout the developing and adult brain and are mutated in some individuals with autism spectrum disorder (ASD). However, how topoisomerases are mechanistically connected to ASD is unknown. Here we find that topotecan, a topoisomerase 1 (TOP1) inhibitor, dose-dependently reduces the expression of extremely long genes in mouse and human neurons, including nearly all genes that are longer than 200 kilobases. Expression of long genes is also reduced after knockdown of Top1 or Top2b in neurons, highlighting that both enzymes are required for full expression of long genes. By mapping RNA polymerase II density genome-wide in neurons, we found that this length-dependent effect on gene expression was due to impaired transcription elongation. Interestingly, many high-confidence ASD candidate genes are exceptionally long and were reduced in expression after TOP1 inhibition. Our findings suggest that chemicals and genetic mutations that impair topoisomerases could commonly contribute to ASD and other neurodevelopmental disorders.
Figures

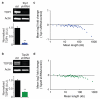
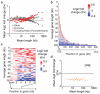
Comment in
-
Autism: A long genetic explanation.Nature. 2013 Sep 5;501(7465):36-7. doi: 10.1038/nature12553. Epub 2013 Aug 28. Nature. 2013. PMID: 23995684 No abstract available.
-
Bias towards large genes in autism.Nature. 2014 Aug 7;512(7512):E1-2. doi: 10.1038/nature13583. Nature. 2014. PMID: 25100484 No abstract available.
-
Zylka et al. reply.Nature. 2014 Aug 7;512(7512):E2. doi: 10.1038/nature13584. Nature. 2014. PMID: 25100485 Free PMC article. No abstract available.
Similar articles
-
Topoisomerase 1 Regulates Gene Expression in Neurons through Cleavage Complex-Dependent and -Independent Mechanisms.PLoS One. 2016 May 27;11(5):e0156439. doi: 10.1371/journal.pone.0156439. eCollection 2016. PLoS One. 2016. PMID: 27231886 Free PMC article.
-
Topoisomerase II binds nucleosome-free DNA and acts redundantly with topoisomerase I to enhance recruitment of RNA Pol II in budding yeast.Proc Natl Acad Sci U S A. 2011 Aug 2;108(31):12693-8. doi: 10.1073/pnas.1106834108. Epub 2011 Jul 19. Proc Natl Acad Sci U S A. 2011. PMID: 21771901 Free PMC article.
-
Sequential topoisomerase targeting and analysis of mechanisms of resistance to topotecan in patients with acute myelogenous leukemia.Anticancer Drugs. 2008 Apr;19(4):411-20. doi: 10.1097/CAD.0b013e3282f5218b. Anticancer Drugs. 2008. PMID: 18454051 Clinical Trial.
-
Topoisomerases interlink genetic network underlying autism.Int J Dev Neurosci. 2015 Dec;47(Pt B):361-8. doi: 10.1016/j.ijdevneu.2015.07.009. Epub 2015 Aug 11. Int J Dev Neurosci. 2015. PMID: 26456455 Review.
-
Tyrosyl-DNA phosphodiesterase inhibitors: Progress and potential.Bioorg Med Chem. 2016 Nov 1;24(21):5017-5027. doi: 10.1016/j.bmc.2016.09.045. Epub 2016 Sep 20. Bioorg Med Chem. 2016. PMID: 27687971 Review.
Cited by
-
Prevalence and mechanisms of somatic deletions in single human neurons during normal aging and in DNA repair disorders.Nat Commun. 2022 Oct 7;13(1):5918. doi: 10.1038/s41467-022-33642-w. Nat Commun. 2022. PMID: 36207339 Free PMC article.
-
IPSC Models of Chromosome 15Q Imprinting Disorders: From Disease Modeling to Therapeutic Strategies.Adv Neurobiol. 2020;25:55-77. doi: 10.1007/978-3-030-45493-7_3. Adv Neurobiol. 2020. PMID: 32578144 Free PMC article.
-
Topoisomerases I and II facilitate condensin DC translocation to organize and repress X chromosomes in C. elegans.Mol Cell. 2022 Nov 17;82(22):4202-4217.e5. doi: 10.1016/j.molcel.2022.10.002. Epub 2022 Oct 26. Mol Cell. 2022. PMID: 36302374 Free PMC article.
-
DNA Topoisomerase I differentially modulates R-loops across the human genome.Genome Biol. 2018 Jul 30;19(1):100. doi: 10.1186/s13059-018-1478-1. Genome Biol. 2018. PMID: 30060749 Free PMC article.
-
rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment.Cells. 2019 Nov 5;8(11):1393. doi: 10.3390/cells8111393. Cells. 2019. PMID: 31694324 Free PMC article.
References
-
- Delorme R, et al. Progress toward treatments for synaptic defects in autism. Nat Med. 2013;19:685–694. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
- R01 MH093372/MH/NIMH NIH HHS/United States
- R01 GM101974/GM/NIGMS NIH HHS/United States
- T32HD040127/HD/NICHD NIH HHS/United States
- P30 NS045892/NS/NINDS NIH HHS/United States
- R01MH093372/MH/NIMH NIH HHS/United States
- P30 HD003110/HD/NICHD NIH HHS/United States
- T32 HD040127/HD/NICHD NIH HHS/United States
- P30HD03110/HD/NICHD NIH HHS/United States
- P30NS045892/NS/NINDS NIH HHS/United States
- R01 HD068730/HD/NICHD NIH HHS/United States
- R01GM101974/GM/NIGMS NIH HHS/United States
- R01HD068730/HD/NICHD NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

