Pyrrolysyl-tRNA synthetase variants reveal ancestral aminoacylation function
- PMID: 23994531
- PMCID: PMC3778162
- DOI: 10.1016/j.febslet.2013.08.018
Pyrrolysyl-tRNA synthetase variants reveal ancestral aminoacylation function
Abstract
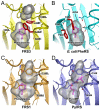
Pyrrolysyl-tRNA synthetase (PylRS) is a class IIc aminoacyl-tRNA synthetase that is related to phenylalanyl-tRNA synthetase (PheRS). Genetic selection provided PylRS variants with a broad range of specificity for diverse non-canonical amino acids (ncAAs). One variant is a specific phenylalanine-incorporating enzyme. Structural models of the PylRSamino acid complex show that the small pocket size and π-interaction play an important role in specific recognition of Phe and the engineered PylRS active site resembles that of Escherichia coli PheRS.
Keywords: Non-canonical amino acid; Phenylalanyl-tRNA synthetase; Pyrrolysyl-tRNA synthetase; Superfolder green fluorescent protein.
Copyright © 2013 Federation of European Biochemical Societies. Published by Elsevier B.V. All rights reserved.
Figures
Similar articles
-
Recognition of non-alpha-amino substrates by pyrrolysyl-tRNA synthetase.J Mol Biol. 2009 Feb 6;385(5):1352-60. doi: 10.1016/j.jmb.2008.11.059. Epub 2008 Dec 11. J Mol Biol. 2009. PMID: 19100747
-
Engineering a Polyspecific Pyrrolysyl-tRNA Synthetase by a High Throughput FACS Screen.Sci Rep. 2019 Aug 19;9(1):11971. doi: 10.1038/s41598-019-48357-0. Sci Rep. 2019. PMID: 31427620 Free PMC article.
-
Aminoacylation of tRNA 2'- or 3'-hydroxyl by phosphoseryl- and pyrrolysyl-tRNA synthetases.FEBS Lett. 2013 Oct 11;587(20):3360-4. doi: 10.1016/j.febslet.2013.08.037. Epub 2013 Sep 8. FEBS Lett. 2013. PMID: 24021645 Free PMC article.
-
Pyrrolysyl-tRNA synthetase: an ordinary enzyme but an outstanding genetic code expansion tool.Biochim Biophys Acta. 2014 Jun;1844(6):1059-70. doi: 10.1016/j.bbapap.2014.03.002. Epub 2014 Mar 12. Biochim Biophys Acta. 2014. PMID: 24631543 Free PMC article. Review.
-
tRNAPyl: Structure, function, and applications.RNA Biol. 2018;15(4-5):441-452. doi: 10.1080/15476286.2017.1356561. Epub 2017 Sep 13. RNA Biol. 2018. PMID: 28837402 Free PMC article. Review.
Cited by
-
Expanding the Scope of Orthogonal Translation with Pyrrolysyl-tRNA Synthetases Dedicated to Aromatic Amino Acids.Molecules. 2020 Sep 25;25(19):4418. doi: 10.3390/molecules25194418. Molecules. 2020. PMID: 32992991 Free PMC article.
-
In Vivo Biosynthesis of a β-Amino Acid-Containing Protein.J Am Chem Soc. 2016 Apr 27;138(16):5194-7. doi: 10.1021/jacs.6b01023. Epub 2016 Apr 18. J Am Chem Soc. 2016. PMID: 27086674 Free PMC article.
-
Expanding the substrate scope of pyrrolysyl-transfer RNA synthetase enzymes to include non-α-amino acids in vitro and in vivo.Nat Chem. 2023 Jul;15(7):960-971. doi: 10.1038/s41557-023-01224-y. Epub 2023 Jun 1. Nat Chem. 2023. PMID: 37264106 Free PMC article.
-
Efficient Reassignment of a Frequent Serine Codon in Wild-Type Escherichia coli.ACS Synth Biol. 2016 Feb 19;5(2):163-71. doi: 10.1021/acssynbio.5b00197. Epub 2015 Nov 20. ACS Synth Biol. 2016. PMID: 26544153 Free PMC article.
-
Rational design of the genetic code expansion toolkit for in vivo encoding of D-amino acids.Front Genet. 2023 Oct 13;14:1277489. doi: 10.3389/fgene.2023.1277489. eCollection 2023. Front Genet. 2023. PMID: 37904728 Free PMC article.
References
-
- Wang YS, Russell WK, Wang Z, Wan W, Dodd LE, Pai PJ, Russell DH, Liu WR. The de novo engineering of pyrrolysyl-tRNA synthetase for genetic incorporation of L-phenylalanine and its derivatives. Mol Biosyst. 2011;7:714–717. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

