H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction
- PMID: 23990607
- PMCID: PMC3847775
- DOI: 10.1101/gr.155028.113
H4K16 acetylation marks active genes and enhancers of embryonic stem cells, but does not alter chromatin compaction
Abstract
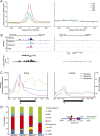
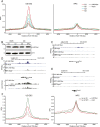
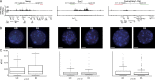
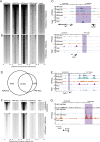
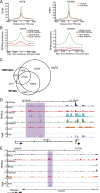
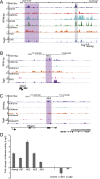
Compared with histone H3, acetylation of H4 tails has not been well studied, especially in mammalian cells. Yet, H4K16 acetylation is of particular interest because of its ability to decompact nucleosomes in vitro and its involvement in dosage compensation in flies. Here we show that, surprisingly, loss of H4K16 acetylation does not alter higher-order chromatin compaction in vivo in mouse embryonic stem cells (ESCs). As well as peaks of acetylated H4K16 and KAT8 histone acetyltransferase at the transcription start sites of expressed genes, we report that acetylation of H4K16 is a new marker of active enhancers in ESCs and that some enhancers are marked by H3K4me1, KAT8, and H4K16ac, but not by acetylated H3K27 or EP300, suggesting that they are novel EP300 independent regulatory elements. Our data suggest a broad role for different histone acetylation marks and for different histone acetyltransferases in long-range gene regulation.
Figures
Similar articles
-
A UTX-MLL4-p300 Transcriptional Regulatory Network Coordinately Shapes Active Enhancer Landscapes for Eliciting Transcription.Mol Cell. 2017 Jul 20;67(2):308-321.e6. doi: 10.1016/j.molcel.2017.06.028. Mol Cell. 2017. PMID: 28732206 Free PMC article.
-
A unique chromatin signature uncovers early developmental enhancers in humans.Nature. 2011 Feb 10;470(7333):279-83. doi: 10.1038/nature09692. Epub 2010 Dec 15. Nature. 2011. PMID: 21160473 Free PMC article.
-
Cross-talk between the H3K36me3 and H4K16ac histone epigenetic marks in DNA double-strand break repair.J Biol Chem. 2017 Jul 14;292(28):11951-11959. doi: 10.1074/jbc.M117.788224. Epub 2017 May 25. J Biol Chem. 2017. PMID: 28546430 Free PMC article.
-
KAT8 beyond Acetylation: A Survey of Its Epigenetic Regulation, Genetic Variability, and Implications for Human Health.Genes (Basel). 2024 May 17;15(5):639. doi: 10.3390/genes15050639. Genes (Basel). 2024. PMID: 38790268 Free PMC article. Review.
-
Nuclear matrix, dynamic histone acetylation and transcriptionally active chromatin.Mol Biol Rep. 1997 Aug;24(3):197-207. doi: 10.1023/a:1006811817247. Mol Biol Rep. 1997. PMID: 9291093 Review.
Cited by
-
The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells.Cell Stem Cell. 2015 Feb 5;16(2):171-83. doi: 10.1016/j.stem.2014.12.004. Epub 2015 Jan 15. Cell Stem Cell. 2015. PMID: 25600643 Free PMC article.
-
Modulation of H4K16Ac levels reduces pro-fibrotic gene expression and mitigates lung fibrosis in aged mice.Theranostics. 2022 Jan 1;12(2):530-541. doi: 10.7150/thno.62760. eCollection 2022. Theranostics. 2022. PMID: 34976199 Free PMC article.
-
Metabolism and acetylation in innate immune cell function and fate.Semin Immunol. 2016 Oct;28(5):408-416. doi: 10.1016/j.smim.2016.10.003. Epub 2016 Oct 27. Semin Immunol. 2016. PMID: 28340958 Free PMC article. Review.
-
Depletion of Histone Demethylase Jarid1A Resulting in Histone Hyperacetylation and Radiation Sensitivity Does Not Affect DNA Double-Strand Break Repair.PLoS One. 2016 Jun 2;11(6):e0156599. doi: 10.1371/journal.pone.0156599. eCollection 2016. PLoS One. 2016. PMID: 27253695 Free PMC article.
-
Separation and Characterization of Endogenous Nucleosomes by Native Capillary Zone Electrophoresis-Top-Down Mass Spectrometry.Anal Chem. 2021 Mar 30;93(12):5151-5160. doi: 10.1021/acs.analchem.0c04975. Epub 2021 Mar 22. Anal Chem. 2021. PMID: 33749242 Free PMC article.
References
-
- Bone JR, Lavender J, Richman R, Palmer MJ, Turner BM, Kuroda MI 1994. Acetylated histone H4 on the male X chromosome is associated with dosage compensation in Drosophila. Genes Dev 8: 96–104 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
