Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1
- PMID: 23963452
- PMCID: PMC3795251
- DOI: 10.1074/jbc.M113.487090
Eukaryotic release factor 3 is required for multiple turnovers of peptide release catalysis by eukaryotic release factor 1
Abstract
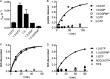
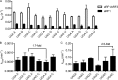
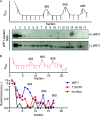
Eukaryotic peptide release factor 3 (eRF3) is a conserved, essential gene in eukaryotes implicated in translation termination. We have systematically measured the contribution of eRF3 to the rates of peptide release with both saturating and limiting levels of eukaryotic release factor 1 (eRF1). Although eRF3 modestly stimulates the absolute rate of peptide release (∼5-fold), it strongly increases the rate of peptide release when eRF1 is limiting (>20-fold). This effect was generalizable across all stop codons and in a variety of contexts. Further investigation revealed that eRF1 remains associated with ribosomal complexes after peptide release and subunit dissociation and that eRF3 promotes the dissociation of eRF1 from these post-termination complexes. These data are consistent with models where eRF3 principally affects binding interactions between eRF1 and the ribosome, either prior to or subsequent to peptide release. A role for eRF3 as an escort for eRF1 into its fully accommodated state is easily reconciled with its close sequence similarity to the translational GTPase EFTu.
Keywords: Protein Synthesis; Ribosomes; Translation; Translation Control; Translation Release Factors; Translational Control.
Figures


Similar articles
-
GTP hydrolysis by eRF3 facilitates stop codon decoding during eukaryotic translation termination.Mol Cell Biol. 2004 Sep;24(17):7769-78. doi: 10.1128/MCB.24.17.7769-7778.2004. Mol Cell Biol. 2004. PMID: 15314182 Free PMC article.
-
Translation termination depends on the sequential ribosomal entry of eRF1 and eRF3.Nucleic Acids Res. 2019 May 21;47(9):4798-4813. doi: 10.1093/nar/gkz177. Nucleic Acids Res. 2019. PMID: 30873535 Free PMC article.
-
N-terminal region of Saccharomyces cerevisiae eRF3 is essential for the functioning of the eRF1/eRF3 complex beyond translation termination.BMC Mol Biol. 2006 Oct 11;7:34. doi: 10.1186/1471-2199-7-34. BMC Mol Biol. 2006. PMID: 17034622 Free PMC article.
-
Genetic reprogramming by retroviruses: enhanced suppression of translational termination.Cell Cycle. 2004 Feb;3(2):123-5. Cell Cycle. 2004. PMID: 14712070 Review.
-
Translation termination and its regulation in eukaryotes: recent insights provided by studies in yeast.Biochemistry (Mosc). 1999 Dec;64(12):1360-6. Biochemistry (Mosc). 1999. PMID: 10648959 Review.
Cited by
-
Oxidation and alkylation stresses activate ribosome-quality control.Nat Commun. 2019 Dec 9;10(1):5611. doi: 10.1038/s41467-019-13579-3. Nat Commun. 2019. PMID: 31819057 Free PMC article.
-
Interaction between the poly(A)-binding protein Pab1 and the eukaryotic release factor eRF3 regulates translation termination but not mRNA decay in Saccharomyces cerevisiae.RNA. 2015 Jan;21(1):124-34. doi: 10.1261/rna.047282.114. Epub 2014 Nov 19. RNA. 2015. PMID: 25411355 Free PMC article.
-
Mechanisms that ensure speed and fidelity in eukaryotic translation termination.Science. 2021 Aug 20;373(6557):876-882. doi: 10.1126/science.abi7801. Science. 2021. PMID: 34413231 Free PMC article.
-
Ribosome quality control antagonizes the activation of the integrated stress response on colliding ribosomes.Mol Cell. 2021 Feb 4;81(3):614-628.e4. doi: 10.1016/j.molcel.2020.11.033. Epub 2020 Dec 17. Mol Cell. 2021. PMID: 33338396 Free PMC article.
-
Ribosome-based quality control of mRNA and nascent peptides.Wiley Interdiscip Rev RNA. 2017 Jan;8(1):10.1002/wrna.1366. doi: 10.1002/wrna.1366. Epub 2016 May 18. Wiley Interdiscip Rev RNA. 2017. PMID: 27193249 Free PMC article. Review.
References
-
- Frolova L. Y., Tsivkovskii R. Y., Sivolobova G. F., Oparina N. Y., Serpinsky O. I., Blinov V. M., Tatkov S. I., Kisselev L. L. (1999) Mutations in the highly conserved GGQ motif of class 1 polypeptide release factors abolish ability of human eRF1 to trigger peptidyl-tRNA hydrolysis. RNA 5, 1014–1020 - PMC - PubMed
-
- Song H., Mugnier P., Das A. K., Webb H. M., Evans D. R., Tuite M. F., Hemmings B. A., Barford D. (2000) The crystal structure of human eukaryotic release factor eRF1–mechanism of stop codon recognition and peptidyl-tRNA hydrolysis. Cell 100, 311–321 - PubMed
-
- Frolova L., Le Goff X., Rasmussen H. H., Cheperegin S., Drugeon G., Kress M., Arman I., Haenni A. L., Celis J. E., Philippe M. (1994) A highly conserved eukaryotic protein family possessing properties of polypeptide chain release factor. Nature 372, 701–703 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

