A role for MeCP2 in switching gene activity via chromatin unfolding and HP1γ displacement
- PMID: 23935992
- PMCID: PMC3720725
- DOI: 10.1371/journal.pone.0069347
A role for MeCP2 in switching gene activity via chromatin unfolding and HP1γ displacement
Abstract
Methyl-CpG-binding protein 2 (MeCP2) is generally considered to act as a transcriptional repressor, whereas recent studies suggest that MeCP2 is also involved in transcription activation. To gain insight into this dual function of MeCP2, we assessed the impact of MeCP2 on higher-order chromatin structure in living cells using mammalian cell systems harbouring a lactose operator and reporter gene-containing chromosomal domain to assess the effect of lactose repressor-tagged MeCP2 (and separate MeCP2 domains) binding in living cells. Our data reveal that targeted binding of MeCP2 elicits extensive chromatin unfolding. MeCP2-induced chromatin unfolding is triggered independently of the methyl-cytosine-binding domain. Interestingly, MeCP2 binding triggers the loss of HP1γ at the chromosomal domain and an increased HP1γ mobility, which is not observed for HP1α and HP1β. Surprisingly, MeCP2-induced chromatin unfolding is not associated with transcriptional activation. Our study suggests a novel role for MeCP2 in reorganizing chromatin to facilitate a switch in gene activity.
Conflict of interest statement
Figures
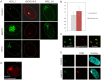
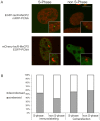
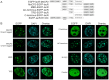
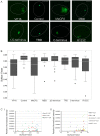
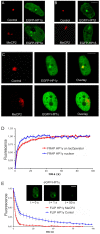
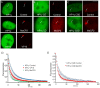
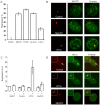
Similar articles
-
MeCP2 is a transcriptional repressor with abundant binding sites in genomic chromatin.Cell. 1997 Feb 21;88(4):471-81. doi: 10.1016/s0092-8674(00)81887-5. Cell. 1997. PMID: 9038338
-
Neuronal MeCP2 is expressed at near histone-octamer levels and globally alters the chromatin state.Mol Cell. 2010 Feb 26;37(4):457-68. doi: 10.1016/j.molcel.2010.01.030. Mol Cell. 2010. PMID: 20188665 Free PMC article.
-
Analysis of protein domains and Rett syndrome mutations indicate that multiple regions influence chromatin-binding dynamics of the chromatin-associated protein MECP2 in vivo.J Cell Sci. 2008 Apr 1;121(Pt 7):1128-37. doi: 10.1242/jcs.016865. Epub 2008 Mar 11. J Cell Sci. 2008. PMID: 18334558 Free PMC article.
-
Binding of the Rett syndrome protein, MeCP2, to methylated and unmethylated DNA and chromatin.IUBMB Life. 2010 Oct;62(10):732-8. doi: 10.1002/iub.386. IUBMB Life. 2010. PMID: 21031501 Free PMC article. Review.
-
Recent advances in MeCP2 structure and function.Biochem Cell Biol. 2009 Feb;87(1):219-27. doi: 10.1139/O08-115. Biochem Cell Biol. 2009. PMID: 19234536 Free PMC article. Review.
Cited by
-
Interplay of LIS1 and MeCP2: Interactions and Implications With the Neurodevelopmental Disorders Lissencephaly and Rett Syndrome.Front Cell Neurosci. 2019 Aug 14;13:370. doi: 10.3389/fncel.2019.00370. eCollection 2019. Front Cell Neurosci. 2019. PMID: 31474834 Free PMC article.
-
MeCP2 Binding Cooperativity Inhibits DNA Modification-Specific Recognition.Biochemistry. 2016 Aug 9;55(31):4275-85. doi: 10.1021/acs.biochem.6b00451. Epub 2016 Jul 28. Biochemistry. 2016. PMID: 27420643 Free PMC article.
-
Maternal Experience-Dependent Cortical Plasticity in Mice Is Circuit- and Stimulus-Specific and Requires MECP2.J Neurosci. 2020 Feb 12;40(7):1514-1526. doi: 10.1523/JNEUROSCI.1964-19.2019. Epub 2020 Jan 7. J Neurosci. 2020. PMID: 31911459 Free PMC article.
-
Osteoblast function and bone histomorphometry in a murine model of Rett syndrome.Bone. 2015 Jul;76:23-30. doi: 10.1016/j.bone.2015.01.024. Epub 2015 Mar 10. Bone. 2015. PMID: 25769649 Free PMC article.
-
The Cell Adhesion Molecule L1 Interacts with Methyl CpG Binding Protein 2 via Its Intracellular Domain.Int J Mol Sci. 2022 Mar 24;23(7):3554. doi: 10.3390/ijms23073554. Int J Mol Sci. 2022. PMID: 35408913 Free PMC article.
References
-
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, et al. (2003) The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem 278: 4035–4040. - PubMed
-
- Shahbazian MD, Antalffy B, Armstrong DL, Zoghbi HY (2002) Insight into Rett syndrome: MeCP2 levels display tissue- and cell-specific differences and correlate with neuronal maturation. Hum Mol Genet 11: 115–124. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

