Cell-cycle dependence of transcription dominates noise in gene expression
- PMID: 23935476
- PMCID: PMC3723585
- DOI: 10.1371/journal.pcbi.1003161
Cell-cycle dependence of transcription dominates noise in gene expression
Abstract
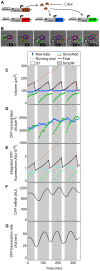
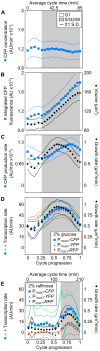
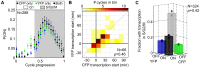
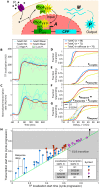
The large variability in mRNA and protein levels found from both static and dynamic measurements in single cells has been largely attributed to random periods of transcription, often occurring in bursts. The cell cycle has a pronounced global role in affecting transcriptional and translational output, but how this influences transcriptional statistics from noisy promoters is unknown and generally ignored by current stochastic models. Here we show that variable transcription from the synthetic tetO promoter in S. cerevisiae is dominated by its dependence on the cell cycle. Real-time measurements of fluorescent protein at high expression levels indicate tetO promoters increase transcription rate ∼2-fold in S/G2/M similar to constitutive genes. At low expression levels, where tetO promoters are thought to generate infrequent bursts of transcription, we observe random pulses of expression restricted to S/G2/M, which are correlated between homologous promoters present in the same cell. The analysis of static, single-cell mRNA measurements at different points along the cell cycle corroborates these findings. Our results demonstrate that highly variable mRNA distributions in yeast are not solely the result of randomly switching between periods of active and inactive gene expression, but instead largely driven by differences in transcriptional activity between G1 and S/G2/M.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

Comment in
-
Gene expression: a cycle route to transcriptional noise.Nat Rev Genet. 2013 Sep;14(9):596. doi: 10.1038/nrg3558. Epub 2013 Aug 13. Nat Rev Genet. 2013. PMID: 23938369 No abstract available.
Similar articles
-
Quantifying how post-transcriptional noise and gene copy number variation bias transcriptional parameter inference from mRNA distributions.Elife. 2022 Oct 17;11:e82493. doi: 10.7554/eLife.82493. Elife. 2022. PMID: 36250630 Free PMC article.
-
Dynamic analysis of stochastic transcription cycles.PLoS Biol. 2011 Apr;9(4):e1000607. doi: 10.1371/journal.pbio.1000607. Epub 2011 Apr 12. PLoS Biol. 2011. PMID: 21532732 Free PMC article.
-
Intercellular Variability in Protein Levels from Stochastic Expression and Noisy Cell Cycle Processes.PLoS Comput Biol. 2016 Aug 18;12(8):e1004972. doi: 10.1371/journal.pcbi.1004972. eCollection 2016 Aug. PLoS Comput Biol. 2016. PMID: 27536771 Free PMC article.
-
Cell cycle-dependent transcription in yeast: promoters, transcription factors, and transcriptomes.Oncogene. 2005 Apr 18;24(17):2746-55. doi: 10.1038/sj.onc.1208606. Oncogene. 2005. PMID: 15838511 Review.
-
Cell cycle regulated transcription in yeast.Curr Opin Cell Biol. 1994 Jun;6(3):451-9. doi: 10.1016/0955-0674(94)90039-6. Curr Opin Cell Biol. 1994. PMID: 7917338 Review.
Cited by
-
A geometric analysis of fast-slow models for stochastic gene expression.J Math Biol. 2016 Jan;72(1-2):87-122. doi: 10.1007/s00285-015-0876-1. Epub 2015 Apr 2. J Math Biol. 2016. PMID: 25833185
-
Quantifying how post-transcriptional noise and gene copy number variation bias transcriptional parameter inference from mRNA distributions.Elife. 2022 Oct 17;11:e82493. doi: 10.7554/eLife.82493. Elife. 2022. PMID: 36250630 Free PMC article.
-
Revealing the vectors of cellular identity with single-cell genomics.Nat Biotechnol. 2016 Nov 8;34(11):1145-1160. doi: 10.1038/nbt.3711. Nat Biotechnol. 2016. PMID: 27824854 Free PMC article. Review.
-
Pathway dynamics can delineate the sources of transcriptional noise in gene expression.Elife. 2021 Oct 12;10:e69324. doi: 10.7554/eLife.69324. Elife. 2021. PMID: 34636320 Free PMC article.
-
Quantifying and correcting bias in transcriptional parameter inference from single-cell data.Biophys J. 2024 Jan 2;123(1):4-30. doi: 10.1016/j.bpj.2023.10.021. Epub 2023 Oct 27. Biophys J. 2024. PMID: 37885177 Free PMC article.
References
-
- To TL, Maheshri N (2010) Noise can induce bimodality in positive transcriptional feedback loops without bistability. Science 327: 1142–1145. - PubMed
-
- Newman JR, Ghaemmaghami S, Ihmels J, Breslow DK, Noble M, et al. (2006) Single-cell proteomic analysis of S. cerevisiae reveals the architecture of biological noise. Nature 441: 840–846. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

