The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation
- PMID: 23926099
- PMCID: PMC3784725
- DOI: 10.1074/jbc.M113.499640
The relaxin receptor (RXFP1) utilizes hydrophobic moieties on a signaling surface of its N-terminal low density lipoprotein class A module to mediate receptor activation
Abstract
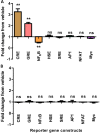
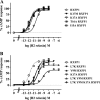
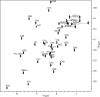
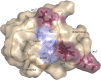
The peptide hormone relaxin is showing potential as a treatment for acute heart failure. Although it is known that relaxin mediates its actions through the G protein-coupled receptor relaxin family peptide receptor 1 (RXFP1), little is known about the molecular mechanisms by which relaxin binding results in receptor activation. Previous studies have highlighted that the unique N-terminal low density lipoprotein class A (LDLa) module of RXFP1 is essential for receptor activation, and it has been hypothesized that this module is the true "ligand" of the receptor that directs the conformational changes necessary for G protein coupling. In this study, we confirmed that an RXFP1 receptor lacking the LDLa module binds ligand normally but cannot signal through any characterized G protein-coupled receptor signaling pathway. Furthermore, we comprehensively examined the contributions of amino acids in the LDLa module to RXFP1 activity using both gain-of-function and loss-of-function mutational analysis together with NMR structural analysis of recombinant LDLa modules. Gain-of-function studies with an inactive RXFP1 chimera containing the LDLa module of the human LDL receptor (LB2) demonstrated two key N-terminal regions of the module that were able to rescue receptor signaling. Loss-of-function mutations of residues in these regions demonstrated that Leu-7, Tyr-9, and Lys-17 all contributed to the ability of the LDLa module to drive receptor activation, and judicious amino acid substitutions suggested this involves hydrophobic interactions. Our results demonstrate that these key residues contribute to interactions driving the active receptor conformation, providing further evidence of a unique mode of G protein-coupled receptor activation.
Keywords: G Protein-coupled Receptors (GPCR); NMR; Peptide Hormones; Peptides; Protein Structure; RXFP1; Relaxin.
Figures




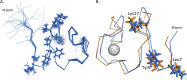
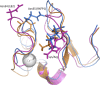
Similar articles
-
The NMR solution structure of the relaxin (RXFP1) receptor lipoprotein receptor class A module and identification of key residues in the N-terminal region of the module that mediate receptor activation.J Biol Chem. 2007 Feb 9;282(6):4172-84. doi: 10.1074/jbc.M609526200. Epub 2006 Dec 4. J Biol Chem. 2007. PMID: 17148455
-
The complex binding mode of the peptide hormone H2 relaxin to its receptor RXFP1.Nat Commun. 2016 Apr 18;7:11344. doi: 10.1038/ncomms11344. Nat Commun. 2016. PMID: 27088579 Free PMC article.
-
Investigation of interactions at the extracellular loops of the relaxin family peptide receptor 1 (RXFP1).J Biol Chem. 2014 Dec 12;289(50):34938-52. doi: 10.1074/jbc.M114.600882. Epub 2014 Oct 28. J Biol Chem. 2014. PMID: 25352603 Free PMC article.
-
Membrane receptors: structure and function of the relaxin family peptide receptors.Mol Cell Endocrinol. 2010 May 14;320(1-2):1-15. doi: 10.1016/j.mce.2010.02.003. Epub 2010 Feb 6. Mol Cell Endocrinol. 2010. PMID: 20138959 Review.
-
Relaxin family peptide receptors--former orphans reunite with their parent ligands to activate multiple signalling pathways.Br J Pharmacol. 2007 Mar;150(6):677-91. doi: 10.1038/sj.bjp.0707140. Epub 2007 Feb 12. Br J Pharmacol. 2007. PMID: 17293890 Free PMC article. Review.
Cited by
-
Expression and Characterization of Relaxin Family Peptide Receptor 1 Variants.Front Pharmacol. 2022 Jan 28;12:826112. doi: 10.3389/fphar.2021.826112. eCollection 2021. Front Pharmacol. 2022. PMID: 35153771 Free PMC article.
-
Distinct activation modes of the Relaxin Family Peptide Receptor 2 in response to insulin-like peptide 3 and relaxin.Sci Rep. 2017 Jun 12;7(1):3294. doi: 10.1038/s41598-017-03638-4. Sci Rep. 2017. PMID: 28607406 Free PMC article.
-
In search of a small molecule agonist of the relaxin receptor RXFP1 for the treatment of liver fibrosis.Sci Rep. 2017 Sep 7;7(1):10806. doi: 10.1038/s41598-017-10521-9. Sci Rep. 2017. PMID: 28883402 Free PMC article.
-
Chimeric RXFP1 and RXFP2 Receptors Highlight the Similar Mechanism of Activation Utilizing Their N-Terminal Low-Density Lipoprotein Class A Modules.Front Endocrinol (Lausanne). 2013 Nov 11;4:171. doi: 10.3389/fendo.2013.00171. eCollection 2013. Front Endocrinol (Lausanne). 2013. PMID: 24273532 Free PMC article.
-
In a Class of Their Own - RXFP1 and RXFP2 are Unique Members of the LGR Family.Front Endocrinol (Lausanne). 2015 Sep 7;6:137. doi: 10.3389/fendo.2015.00137. eCollection 2015. Front Endocrinol (Lausanne). 2015. PMID: 26441827 Free PMC article. Review.
References
-
- Bathgate R. A., Samuel C. S., Burazin T. C., Layfield S., Claasz A. A., Reytomas I. G., Dawson N. F., Zhao C., Bond C., Summers R. J., Parry L. J., Wade J. D., Tregear G. W. (2002) Human relaxin gene 3 (H3) and the equivalent mouse relaxin (M3) gene. Novel members of the relaxin peptide family. J. Biol. Chem. 277, 1148–1157 - PubMed
-
- Hsu S. Y. (2003) New insights into the evolution of the relaxin-LGR signaling system. Trends Endocrinol. Metab. 14, 303–309 - PubMed
-
- Evans B. A., Fu P., Tregear G. W. (1994) Characterization of two relaxin genes in the chimpanzee. J. Endocrinol. 140, 385–392 - PubMed
-
- Bathgate R. A., Halls M. L., van der Westhuizen E. T., Callander G. E., Kocan M., Summers R. J. (2013) Relaxin family peptides and their receptors. Physiol. Rev. 93, 405–480 - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

