Chlamydia trachomatis homotypic inclusion fusion is promoted by host microtubule trafficking
- PMID: 23919807
- PMCID: PMC3750546
- DOI: 10.1186/1471-2180-13-185
Chlamydia trachomatis homotypic inclusion fusion is promoted by host microtubule trafficking
Abstract
Background: The developmental cycle of the obligate intracellular pathogen Chlamydia is dependant on the formation of a unique intracellular niche termed the chlamydial inclusion. The inclusion is a membrane bound vacuole derived from host cytoplasmic membrane and is modified significantly by the insertion of chlamydial proteins. A unique property of the inclusion is its propensity for homotypic fusion. The vast majority of cells infected with multiple chlamydial elementary bodies (EBs) contain only a single mature inclusion. The chlamydial protein IncA is required for fusion, however the host process involved are uncharacterized.
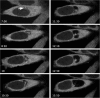
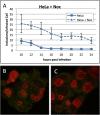
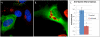
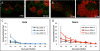
Results: Here, through live imaging studies, we determined that the nascent inclusions clustered tightly at the cell microtubule organizing center (MTOC) where they eventually fused to form a single inclusion. We established that factors involved in trafficking were required for efficient fusion as both disruption of the microtubule network and inhibition of microtubule trafficking reduced the efficiency of fusion. Additionally, fusion occurred at multiple sites in the cell and was delayed when the microtubule minus ends were either no longer anchored at a single MTOC or when a cell possessed multiple MTOCs.
Conclusions: The data presented demonstrates that efficient homotypic fusion requires the inclusions to be in close proximity and that this proximity is dependent on chlamydial microtubule trafficking to the minus ends of microtubules.
Figures
Similar articles
-
Chlamydia trachomatis uses host cell dynein to traffic to the microtubule-organizing center in a p50 dynamitin-independent process.J Cell Sci. 2003 Sep 15;116(Pt 18):3793-802. doi: 10.1242/jcs.00695. Epub 2003 Aug 5. J Cell Sci. 2003. PMID: 12902405
-
Specific chlamydial inclusion membrane proteins associate with active Src family kinases in microdomains that interact with the host microtubule network.Cell Microbiol. 2010 Sep 1;12(9):1235-49. doi: 10.1111/j.1462-5822.2010.01465.x. Epub 2010 Mar 19. Cell Microbiol. 2010. PMID: 20331642 Free PMC article.
-
A meta-analysis of affinity purification-mass spectrometry experimental systems used to identify eukaryotic and chlamydial proteins at the Chlamydia trachomatis inclusion membrane.J Proteomics. 2020 Feb 10;212:103595. doi: 10.1016/j.jprot.2019.103595. Epub 2019 Nov 21. J Proteomics. 2020. PMID: 31760040 Free PMC article.
-
Chlamydia trachomatis and its interaction with the cellular retromer.Int J Med Microbiol. 2018 Jan;308(1):197-205. doi: 10.1016/j.ijmm.2017.10.006. Epub 2017 Oct 26. Int J Med Microbiol. 2018. PMID: 29122514 Review.
-
Got mutants? How advances in chlamydial genetics have furthered the study of effector proteins.Pathog Dis. 2021 Feb 4;79(2):ftaa078. doi: 10.1093/femspd/ftaa078. Pathog Dis. 2021. PMID: 33512479 Free PMC article. Review.
Cited by
-
Chlamydia trachomatis Cell-to-Cell Spread through Tunneling Nanotubes.Microbiol Spectr. 2022 Dec 21;10(6):e0281722. doi: 10.1128/spectrum.02817-22. Epub 2022 Oct 11. Microbiol Spectr. 2022. PMID: 36219107 Free PMC article.
-
Modulation of Host Lipid Pathways by Pathogenic Intracellular Bacteria.Pathogens. 2020 Jul 28;9(8):614. doi: 10.3390/pathogens9080614. Pathogens. 2020. PMID: 32731350 Free PMC article. Review.
-
Chlamydia cell biology and pathogenesis.Nat Rev Microbiol. 2016 Jun;14(6):385-400. doi: 10.1038/nrmicro.2016.30. Epub 2016 Apr 25. Nat Rev Microbiol. 2016. PMID: 27108705 Free PMC article. Review.
-
A Chlamydia effector recruits CEP170 to reprogram host microtubule organization.J Cell Sci. 2015 Sep 15;128(18):3420-34. doi: 10.1242/jcs.169318. Epub 2015 Jul 28. J Cell Sci. 2015. PMID: 26220855 Free PMC article.
-
Taking control: reorganization of the host cytoskeleton by Chlamydia.F1000Res. 2017 Nov 29;6:2058. doi: 10.12688/f1000research.12316.1. eCollection 2017. F1000Res. 2017. PMID: 29225789 Free PMC article. Review.
References
-
- Clifton DR, Fields KA, Grieshaber SS, Dooley CA, Fischer ER, Mead DJ, Carabeo RA, Hackstadt T. A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc Natl Acad Sci USA. 2004;101:10166–10171. doi: 10.1073/pnas.0402829101. - DOI - PMC - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

