Optimization of scarless human stem cell genome editing
- PMID: 23907390
- PMCID: PMC3799423
- DOI: 10.1093/nar/gkt555
Optimization of scarless human stem cell genome editing
Abstract
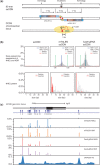
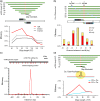
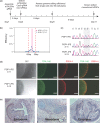
Efficient strategies for precise genome editing in human-induced pluripotent cells (hiPSCs) will enable sophisticated genome engineering for research and clinical purposes. The development of programmable sequence-specific nucleases such as Transcription Activator-Like Effectors Nucleases (TALENs) and Cas9-gRNA allows genetic modifications to be made more efficiently at targeted sites of interest. However, many opportunities remain to optimize these tools and to enlarge their spheres of application. We present several improvements: First, we developed functional re-coded TALEs (reTALEs), which not only enable simple one-pot TALE synthesis but also allow TALE-based applications to be performed using lentiviral vectors. We then compared genome-editing efficiencies in hiPSCs mediated by 15 pairs of reTALENs and Cas9-gRNA targeting CCR5 and optimized ssODN design in conjunction with both methods for introducing specific mutations. We found Cas9-gRNA achieved 7-8× higher non-homologous end joining efficiencies (3%) than reTALENs (0.4%) and moderately superior homology-directed repair efficiencies (1.0 versus 0.6%) when combined with ssODN donors in hiPSCs. Using the optimal design, we demonstrated a streamlined process to generated seamlessly genome corrected hiPSCs within 3 weeks.
Figures

Similar articles
-
Efficient and allele-specific genome editing of disease loci in human iPSCs.Mol Ther. 2015 Mar;23(3):570-7. doi: 10.1038/mt.2014.226. Epub 2014 Nov 24. Mol Ther. 2015. PMID: 25418680 Free PMC article.
-
Genome Editing in Human Induced Pluripotent Stem Cells (hiPSCs).Methods Mol Biol. 2021;2320:235-245. doi: 10.1007/978-1-0716-1484-6_21. Methods Mol Biol. 2021. PMID: 34302662
-
Efficient, footprint-free human iPSC genome editing by consolidation of Cas9/CRISPR and piggyBac technologies.Nat Protoc. 2017 Jan;12(1):88-103. doi: 10.1038/nprot.2016.152. Epub 2016 Dec 8. Nat Protoc. 2017. PMID: 27929521 Free PMC article.
-
TALE: a tale of genome editing.Prog Biophys Mol Biol. 2014 Jan;114(1):25-32. doi: 10.1016/j.pbiomolbio.2013.11.006. Epub 2013 Nov 27. Prog Biophys Mol Biol. 2014. PMID: 24291598 Review.
-
Origins of Programmable Nucleases for Genome Engineering.J Mol Biol. 2016 Feb 27;428(5 Pt B):963-89. doi: 10.1016/j.jmb.2015.10.014. Epub 2015 Oct 23. J Mol Biol. 2016. PMID: 26506267 Free PMC article. Review.
Cited by
-
Genetic rearrangements of variable di-residue (RVD)-containing repeat arrays in a baculoviral TALEN system.Mol Ther Methods Clin Dev. 2014 Oct 29;1:14050. doi: 10.1038/mtm.2014.50. eCollection 2014. Mol Ther Methods Clin Dev. 2014. PMID: 26015987 Free PMC article.
-
Analysis of gene repair tracts from Cas9/gRNA double-stranded breaks in the human CFTR gene.Sci Rep. 2016 Aug 25;6:32230. doi: 10.1038/srep32230. Sci Rep. 2016. PMID: 27557525 Free PMC article.
-
Identification and single-base gene-editing functional validation of a cis-EPO variant as a genetic predictor for EPO-increasing therapies.Am J Hum Genet. 2022 Sep 1;109(9):1638-1652. doi: 10.1016/j.ajhg.2022.08.004. Am J Hum Genet. 2022. PMID: 36055212 Free PMC article.
-
Utilization of TALEN and CRISPR/Cas9 technologies for gene targeting and modification.Exp Biol Med (Maywood). 2015 Aug;240(8):1065-70. doi: 10.1177/1535370215584932. Epub 2015 May 7. Exp Biol Med (Maywood). 2015. PMID: 25956682 Free PMC article. Review.
-
In search of an ideal template for therapeutic genome editing: A review of current developments for structure optimization.Front Genome Ed. 2023 Feb 22;5:1068637. doi: 10.3389/fgeed.2023.1068637. eCollection 2023. Front Genome Ed. 2023. PMID: 36911237 Free PMC article. Review.
References
-
- Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu. Rev. Genet. 2011;45:247–271. - PubMed
-
- Urnov FD, Miller JC, Lee Y-L, Beausejour CM, Rock JM, Augustus S, Jamieson AC, Porteus MH, Gregory PD, Holmes MC. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

