Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila
- PMID: 23906716
- PMCID: PMC3721279
- DOI: 10.1101/cshperspect.a017780
Position-effect variegation, heterochromatin formation, and gene silencing in Drosophila
Abstract
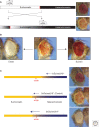
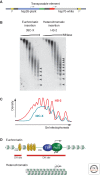
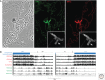
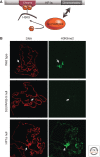
Position-effect variegation (PEV) results when a gene normally in euchromatin is juxtaposed with heterochromatin by rearrangement or transposition. When heterochromatin packaging spreads across the heterochromatin/euchromatin border, it causes transcriptional silencing in a stochastic pattern. PEV is intensely studied in Drosophila using the white gene. Screens for dominant mutations that suppress or enhance white variegation have identified many conserved epigenetic factors, including the histone H3 lysine 9 methyltransferase SU(VAR)3-9. Heterochromatin protein HP1a binds H3K9me2/3 and interacts with SU(VAR)3-9, creating a core memory system. Genetic, molecular, and biochemical analysis of PEV in Drosophila has contributed many key findings concerning establishment and maintenance of heterochromatin with concomitant gene silencing.
Figures


Similar articles
-
Live analysis of position-effect variegation in Drosophila reveals different modes of action for HP1a and Su(var)3-9.Proc Natl Acad Sci U S A. 2022 Jun 21;119(25):e2118796119. doi: 10.1073/pnas.2118796119. Epub 2022 Jun 15. Proc Natl Acad Sci U S A. 2022. PMID: 35704756 Free PMC article.
-
[Correlation on a cellular level of gene transcriptional silencing and heterochromatin compartment dragging in case of PEV-producing eu-heterochromatin rearrangement in Drosophila melanogaster].Mol Biol (Mosk). 2013 Mar-Apr;47(2):286-91. doi: 10.7868/s0026898413020080. Mol Biol (Mosk). 2013. PMID: 23808163 Russian.
-
Histone modification and the control of heterochromatic gene silencing in Drosophila.Chromosome Res. 2006;14(4):377-92. doi: 10.1007/s10577-006-1066-1. Chromosome Res. 2006. PMID: 16821134 Review.
-
Mutations in CG8878, a novel putative protein kinase, enhance P element dependent silencing (PDS) and position effect variegation (PEV) in Drosophila melanogaster.PLoS One. 2014 Mar 10;9(3):e71695. doi: 10.1371/journal.pone.0071695. eCollection 2014. PLoS One. 2014. PMID: 24614804 Free PMC article.
-
Position-effect variegation and the genetic dissection of chromatin regulation in Drosophila.Semin Cell Dev Biol. 2003 Feb;14(1):67-75. doi: 10.1016/s1084-9521(02)00138-6. Semin Cell Dev Biol. 2003. PMID: 12524009 Review.
Cited by
-
H3K9 methylation regulates heterochromatin silencing through incoherent feedforward loops.Sci Adv. 2024 Jun 28;10(26):eadn4149. doi: 10.1126/sciadv.adn4149. Epub 2024 Jun 26. Sci Adv. 2024. PMID: 38924413 Free PMC article.
-
Zinc Metalloproteins in Epigenetics and Their Crosstalk.Life (Basel). 2021 Feb 26;11(3):186. doi: 10.3390/life11030186. Life (Basel). 2021. PMID: 33652690 Free PMC article. Review.
-
Direct interrogation of the role of H3K9 in metazoan heterochromatin function.Genes Dev. 2016 Aug 15;30(16):1866-80. doi: 10.1101/gad.286278.116. Epub 2016 Aug 26. Genes Dev. 2016. PMID: 27566777 Free PMC article.
-
Genome-Wide Association Study and transcriptome analysis reveals a complex gene network that regulates opsin gene expression and cell fate determination in Drosophila R7 photoreceptor cells.bioRxiv [Preprint]. 2024 Aug 7:2024.08.05.606616. doi: 10.1101/2024.08.05.606616. bioRxiv. 2024. PMID: 39149333 Free PMC article. Preprint.
-
Selective nucleolus organizer inactivation in Arabidopsis is a chromosome position-effect phenomenon.Proc Natl Acad Sci U S A. 2016 Nov 22;113(47):13426-13431. doi: 10.1073/pnas.1608140113. Epub 2016 Nov 7. Proc Natl Acad Sci U S A. 2016. PMID: 27821753 Free PMC article.
References
-
- Ahmad K, Henikoff S 2001. Modulation of a transcription factor counteracts heterochromatin gene silencing in Drosophila. Cell 104: 839–847 - PubMed
-
- Aravin AA, Lagos-Quintana M, Yalcin A, Zavolan M, Marks D, Snyder B, Gaasterland T, Meyer J, Tuschl T 2003. The small RNA profile during Drosophila melanogaster development. Dev Cell 5: 337–350 - PubMed
-
- Ashburner M, Golic KG, Hawley RS 2005. Chromosomes and position effect variegation. In Drosophila: A laboratory handbook, 2nd ed., Chaps. 4, 28 Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
